Coilin participates in the suppression of RNA polymerase I in response to cisplatin-induced DNA damage
- PMID: 21289084
- PMCID: PMC3069010
- DOI: 10.1091/mbc.E10-08-0731
Coilin participates in the suppression of RNA polymerase I in response to cisplatin-induced DNA damage
Abstract
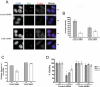
Coilin is a nuclear phosphoprotein that concentrates within Cajal bodies (CBs) and impacts small nuclear ribonucleoprotein (snRNP) biogenesis. Cisplatin and γ-irradiation, which cause distinct types of DNA damage, both trigger the nucleolar accumulation of coilin, and this temporally coincides with the repression of RNA polymerase I (Pol I) activity. Knockdown of endogenous coilin partially overrides the Pol I transcriptional arrest caused by cisplatin, while both ectopically expressed and exogenous coilin accumulate in the nucleolus and suppress rRNA synthesis. In support of this mechanism, we demonstrate that both cisplatin and γ-irradiation induce the colocalization of coilin with RPA-194 (the largest subunit of Pol I), and we further show that coilin can specifically interact with RPA-194 and the key regulator of Pol I activity, upstream binding factor (UBF). Using chromatin immunoprecipitation analysis, we provide evidence that coilin modulates the association of Pol I with ribosomal DNA. Collectively, our data suggest that coilin acts to repress Pol I activity in response to cisplatin-induced DNA damage. Our findings identify a novel and unexpected function for coilin, independent of its role in snRNP biogenesis, establishing a new link between the DNA damage response and the inhibition of rRNA synthesis.
Figures





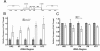
References
-
- Bartek J, Lukas C, Lukas J. Checking on DNA damage in S phase. Nat Rev Mol Cell Biol. 2004;5:792–804. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

