Functional characterization of putative cilia genes by high-content analysis
- PMID: 21289087
- PMCID: PMC3069013
- DOI: 10.1091/mbc.E10-07-0596
Functional characterization of putative cilia genes by high-content analysis
Abstract
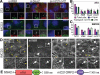
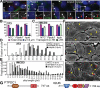
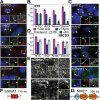
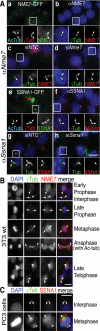
Cilia are microtubule-based protrusions from the cell surface that are involved in a number of essential signaling pathways, yet little is known about many of the proteins that regulate their structure and function. A number of putative cilia genes have been identified by proteomics and comparative sequence analyses, but functional data are lacking for the vast majority. We therefore monitored the effects in three cell lines of small interfering RNA (siRNA) knockdown of 40 of these genes by high-content analysis. We assayed cilia number, length, and transport of two different cargoes (membranous serotonin receptor 6-green fluorescent protein [HTR6-GFP] and the endogenous Hedgehog [Hh] pathway transcription factor Gli3) by immunofluorescence microscopy; and cilia function using a Gli-luciferase Hh signaling assay. Hh signaling was most sensitive to perturbations, with or without visible structural cilia defects. Validated hits include Ssa2 and mC21orf2 with ciliation defects; Ift46 with short cilia; Ptpdc1 and Iqub with elongated cilia; and Arl3, Nme7, and Ssna1 with distinct ciliary transport but not length defects. Our data confirm various ciliary roles for several ciliome proteins and show it is possible to uncouple ciliary cargo transport from cilia formation in vertebrates.
Figures





References
-
- Avidor-Reiss T, Maer AM, Koundakjian E, Polyanovsky A, Keil T, Subramaniam S, Zuker CS. Decoding cilia function: defining specialized genes required for compartmentalized cilia biogenesis. Cell. 2004;117:527–539. - PubMed
-
- Badano JL, Mitsuma N, Beales PL, Katsanis N. The ciliopathies: an emerging class of human genetic disorders. Annu Rev Genomics Hum Genet. 2006;7:125–148. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

