TRIM56 is a virus- and interferon-inducible E3 ubiquitin ligase that restricts pestivirus infection
- PMID: 21289118
- PMCID: PMC3126137
- DOI: 10.1128/JVI.02546-10
TRIM56 is a virus- and interferon-inducible E3 ubiquitin ligase that restricts pestivirus infection
Abstract
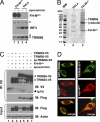
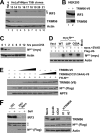
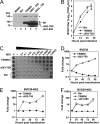
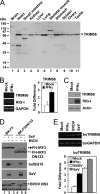
The tripartite motif (TRIM) protein family comprises more than 60 members that have diverse functions in various biological processes. Although a small number of TRIM proteins have been shown to regulate innate immunity, much remains to be learned about the functions of the majority of the TRIM proteins. Here we identify TRIM56 as a cellular protein associated with the N-terminal protease (N(pro)) of bovine viral diarrhea virus (BVDV), a pestiviral interferon antagonist which degrades interferon regulatory factor 3 (IRF3) through the proteasome. We found that TRIM56 was constitutively expressed in most tissues, and its abundance was further upregulated moderately by interferon or virus. The manipulation of TRIM56 abundance did not affect the protein turnover of N(pro) and IRF3. Rather, ectopic expression of TRIM56 substantially impaired, while knockdown of TRIM56 expression greatly enhanced, BVDV replication in cell culture. The antiviral activity of TRIM56 depended on its E3 ubiquitin ligase activity as well as the integrity of its C-terminal region but was not attributed to a general augmentation of the interferon antiviral response. Overexpression of TRIM56 did not inhibit the replication of vesicular stomatitis virus or hepatitis C virus, a virus closely related to BVDV. Together, our data demonstrate that TRIM56 is a novel antiviral host factor that restricts pestivirus infection.
Figures






References
-
- Beard M. R., et al. 1999. An infectious molecular clone of a Japanese genotype 1b hepatitis C virus. Hepatology 30:316–324 - PubMed
-
- Chan M. W., et al. 2005. Hypermethylation of 18S and 28S ribosomal DNAs predicts progression-free survival in patients with ovarian cancer. Clin. Cancer Res. 11:7376–7383 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

