Advances in structure elucidation of small molecules using mass spectrometry
- PMID: 21289855
- PMCID: PMC3015162
- DOI: 10.1007/s12566-010-0015-9
Advances in structure elucidation of small molecules using mass spectrometry
Abstract
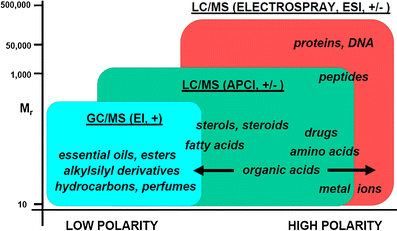
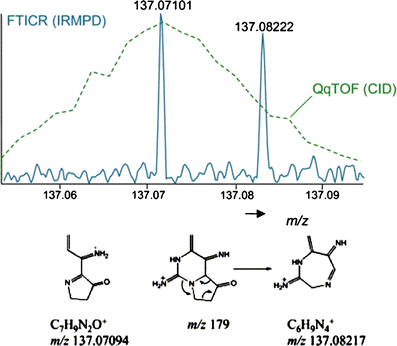
The structural elucidation of small molecules using mass spectrometry plays an important role in modern life sciences and bioanalytical approaches. This review covers different soft and hard ionization techniques and figures of merit for modern mass spectrometers, such as mass resolving power, mass accuracy, isotopic abundance accuracy, accurate mass multiple-stage MS(n) capability, as well as hybrid mass spectrometric and orthogonal chromatographic approaches. The latter part discusses mass spectral data handling strategies, which includes background and noise subtraction, adduct formation and detection, charge state determination, accurate mass measurements, elemental composition determinations, and complex data-dependent setups with ion maps and ion trees. The importance of mass spectral library search algorithms for tandem mass spectra and multiple-stage MS(n) mass spectra as well as mass spectral tree libraries that combine multiple-stage mass spectra are outlined. The successive chapter discusses mass spectral fragmentation pathways, biotransformation reactions and drug metabolism studies, the mass spectral simulation and generation of in silico mass spectra, expert systems for mass spectral interpretation, and the use of computational chemistry to explain gas-phase phenomena. A single chapter discusses data handling for hyphenated approaches including mass spectral deconvolution for clean mass spectra, cheminformatics approaches and structure retention relationships, and retention index predictions for gas and liquid chromatography. The last section reviews the current state of electronic data sharing of mass spectra and discusses the importance of software development for the advancement of structure elucidation of small molecules. ELECTRONIC SUPPLEMENTARY MATERIAL: The online version of this article (doi:10.1007/s12566-010-0015-9) contains supplementary material, which is available to authorized users.
Conflict of interest statement
The authors declare no competing interests.
Figures













References
-
- Brunnée C. The ideal mass analyzer: fact or fiction? Int J Mass Spectrom Ion Processes. 1987;76(2):125–237.
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
