Functional effects of genetic polymorphisms in the N-acetyltransferase 1 coding and 3' untranslated regions
- PMID: 21290563
- PMCID: PMC3252750
- DOI: 10.1002/bdra.20763
Functional effects of genetic polymorphisms in the N-acetyltransferase 1 coding and 3' untranslated regions
Abstract
Background: The functional effects of N-acetyltransferase 1 (NAT1) polymorphisms and haplotypes are poorly understood, compromising the validity of associations reported with diseases, including birth defects and numerous cancers.
Methods: We investigated the effects of genetic polymorphisms within the NAT1 coding region and the 3'-untranslated region (3'-UTR) and their associated haplotypes on N- and O-acetyltransferase catalytic activities, and NAT1 mRNA and protein levels following recombinant expression in COS-1 cells.
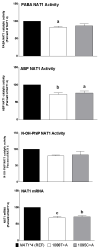
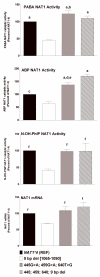
Results: 1088T>A (rs1057126; 3'-UTR) and 1095C>A (rs15561; 3'-UTR) each slightly reduced NAT1 catalytic activity and NAT1 mRNA and protein levels. A 9-bp (TAATAATAA) deletion between nucleotides 1065 and 1090 (3'-UTR) reduced NAT1 catalytic activity and NAT1 mRNA and protein levels. In contrast, a 445G>A (rs4987076; V149I), 459G>A (rs4986990; T153T), and 640T>G (rs4986783; S214A) coding region haplotype present in NAT1*11 increased NAT1 catalytic activity and NAT1 protein, but not NAT1 mRNA levels. A combination of the 9-bp (TAATAATAA) deletion and the 445G>A, 459G>A, and 640T>G coding region haplotypes, both present in NAT1*11, appeared to neutralize the opposing effects on NAT1 protein and catalytic activity, resulting in levels of NAT1 protein and catalytic activity that did not differ significantly from the NAT1*4 reference.
Conclusions: Because 1095C>A (3'-UTR) is the sole polymorphism present in NAT1*3, our data suggest that NAT1*3 is not functionally equivalent to the NAT1*4 reference. Furthermore, our findings provide biologic support for reported associations of 1088T>A and 1095C>A polymorphisms with birth defects.
Copyright © 2011 Wiley-Liss, Inc.
Figures

References
-
- Ambrosone CB, Abrams SM, Gorlewska-Roberts K, Kadlubar FF. Hair dye use, meat intake, and tobacco exposure and presence of carcinogen-DNA adducts in exfoliated breast ductal epithelial cells. Arch Biochem Biophys. 2007;464:169–175. - PubMed
-
- Bell DA, Stephens EA, Castranio T, Umbach DM, Watson M, Deakin M, Elder J, Hendrickse C, Duncan H, Strange RC. Polyadenylation polymorphism in the acetyltransferase 1 gene (NAT1) increases risk of colorectal cancer. Cancer Res. 1995;55:3537–3542. - PubMed
-
- Bruhn C, Brockmoller J, Cascorbi I, Roots I, Borchert HH. Correlation between genotype and phenotype of the human arylamine N-acetyltransferase type 1 (NAT1) Biochem Pharmacol. 1999;58:1759–1764. - PubMed
-
- Carmichael SL, Shaw GM, Yang W, Iovannisci DM, Lammer E. Risk of limb deficiency defects associated with NAT1, NAT2, GSTT1, GSTM1, and NOS3 genetic variants, maternal smoking, and vitamin supplement intake. Am J Med Genet A. 2006;140:1915–1922. - PubMed
-
- Chen B, Zhang WX, Cai WM. The influence of various genotypes on the metabolic activity of NAT2 in a Chinese population. Eur J Clin Pharmacol. 2006;62:355–359. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

