A conserved surface loop in type I dehydroquinate dehydratases positions an active site arginine and functions in substrate binding
- PMID: 21291284
- PMCID: PMC3062685
- DOI: 10.1021/bi102020s
A conserved surface loop in type I dehydroquinate dehydratases positions an active site arginine and functions in substrate binding
Abstract
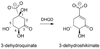
Dehydroquinate dehydratase (DHQD) catalyzes the third step in the biosynthetic shikimate pathway. We present three crystal structures of the Salmonella enterica type I DHQD that address the functionality of a surface loop that is observed to close over the active site following substrate binding. Two wild-type structures with differing loop conformations and kinetic and structural studies of a mutant provide evidence of both direct and indirect mechanisms of involvement of the loop in substrate binding. In addition to allowing amino acid side chains to establish a direct interaction with the substrate, closure of the loop necessitates a conformational change of a key active site arginine, which in turn positions the substrate productively. The absence of DHQD in humans and its essentiality in many pathogenic bacteria make the enzyme a target for the development of nontoxic antimicrobials. The structures and ligand binding insights presented here may inform the design of novel type I DHQD inhibiting molecules.
Figures

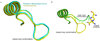
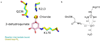
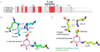
Similar articles
-
Crystal structures of type I dehydroquinate dehydratase in complex with quinate and shikimate suggest a novel mechanism of Schiff base formation.Biochemistry. 2014 Feb 11;53(5):872-80. doi: 10.1021/bi4015506. Epub 2014 Jan 31. Biochemistry. 2014. PMID: 24437575 Free PMC article.
-
Reassessing the type I dehydroquinate dehydratase catalytic triad: kinetic and structural studies of Glu86 mutants.Protein Sci. 2013 Apr;22(4):418-24. doi: 10.1002/pro.2218. Epub 2013 Feb 11. Protein Sci. 2013. PMID: 23341204 Free PMC article.
-
Insights into the mechanism of type I dehydroquinate dehydratases from structures of reaction intermediates.J Biol Chem. 2011 Feb 4;286(5):3531-9. doi: 10.1074/jbc.M110.192831. Epub 2010 Nov 18. J Biol Chem. 2011. PMID: 21087925 Free PMC article.
-
Active site comparisons and catalytic mechanisms of the hot dog superfamily.Chem Rev. 2013 Mar 13;113(3):2182-204. doi: 10.1021/cr300169a. Epub 2012 Dec 3. Chem Rev. 2013. PMID: 23205964 Free PMC article. Review. No abstract available.
-
Nucleotide sugar dehydratases: Structure, mechanism, substrate specificity, and application potential.J Biol Chem. 2022 Apr;298(4):101809. doi: 10.1016/j.jbc.2022.101809. Epub 2022 Mar 7. J Biol Chem. 2022. PMID: 35271853 Free PMC article. Review.
Cited by
-
Discovery of selective inhibitors of the Clostridium difficile dehydroquinate dehydratase.PLoS One. 2014 Feb 21;9(2):e89356. doi: 10.1371/journal.pone.0089356. eCollection 2014. PLoS One. 2014. PMID: 24586713 Free PMC article.
-
Crystal structures of type I dehydroquinate dehydratase in complex with quinate and shikimate suggest a novel mechanism of Schiff base formation.Biochemistry. 2014 Feb 11;53(5):872-80. doi: 10.1021/bi4015506. Epub 2014 Jan 31. Biochemistry. 2014. PMID: 24437575 Free PMC article.
-
Reassessing the type I dehydroquinate dehydratase catalytic triad: kinetic and structural studies of Glu86 mutants.Protein Sci. 2013 Apr;22(4):418-24. doi: 10.1002/pro.2218. Epub 2013 Feb 11. Protein Sci. 2013. PMID: 23341204 Free PMC article.
-
Adherence to Bürgi-Dunitz stereochemical principles requires significant structural rearrangements in Schiff-base formation: insights from transaldolase complexes.Acta Crystallogr D Biol Crystallogr. 2014 Feb;70(Pt 2):544-52. doi: 10.1107/S1399004713030666. Epub 2014 Jan 31. Acta Crystallogr D Biol Crystallogr. 2014. PMID: 24531488 Free PMC article.
-
Identification of polyketide inhibitors targeting 3-dehydroquinate dehydratase in the shikimate pathway of Enterococcus faecalis.PLoS One. 2014 Jul 29;9(7):e103598. doi: 10.1371/journal.pone.0103598. eCollection 2014. PLoS One. 2014. PMID: 25072253 Free PMC article.
References
-
- Bentley R. The shikimate pathway--a metabolic tree with many branches. Crit Rev Biochem Mol Biol. 1990;25:307–384. - PubMed
-
- Benowitz AB, Hoover Jennifer L, Payne David J. Antibacterial Drug Discovery in the Age of Resistance. Microbe. 2010;5:390–396.
-
- Microbe GM, Shah DM. Amino-Acid Biosynthesis Inhibitors as Herbicides. Annu Rev Biochem. 1988;57:627–663. - PubMed
-
- Marques MR, Pereira JH, Oliveira JS, Basso LA, de Azevedo WF, Santos DS, Palma MS. The inhibition of 5-enolpyruvylshikimate-3-phosphate synthase as a model for development of novel antimicrobials. Curr Drug Targets. 2007;8:445–457. - PubMed
-
- Noble M, Sinha Y, Kolupaev A, Demin O, Earnshaw D, Tobin F, West J, Martin JD, Qiu CY, Liu WS, DeWolf WE, Tew D, Goryanin II. The kinetic model of the shikimate pathway as a tool to optimize enzyme assays for high-throughput screening. Biotechnol Bioeng. 2006;95:560–573. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

