Single-molecule studies of RNA polymerase: one singular sensation, every little step it takes
- PMID: 21292158
- PMCID: PMC3056354
- DOI: 10.1016/j.molcel.2011.01.008
Single-molecule studies of RNA polymerase: one singular sensation, every little step it takes
Abstract
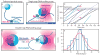
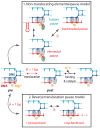
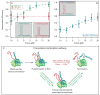
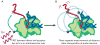
Transcription is the first of many biochemical steps that turn the genetic information found in DNA into the proteins responsible for driving cellular processes. In this review, we highlight certain advantages of single-molecule techniques in the study of prokaryotic and eukaryotic transcription, and the specific ways in which these techniques complement conventional, ensemble-based biochemistry. We focus on recent literature, highlighting examples where single-molecule methods have provided fresh insights into mechanism. We also present recent technological advances and outline future directions in the field.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures

References
-
- Allison LA, Moyle M, Shales M, Ingles CJ. Extensive homology among the largest subunits of eukaryotic and prokaryotic RNA polymerases. Cell. 1985;42:599–610. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

