The base-pairing RNA spot 42 participates in a multioutput feedforward loop to help enact catabolite repression in Escherichia coli
- PMID: 21292161
- PMCID: PMC3072601
- DOI: 10.1016/j.molcel.2010.12.027
The base-pairing RNA spot 42 participates in a multioutput feedforward loop to help enact catabolite repression in Escherichia coli
Abstract
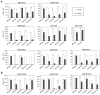
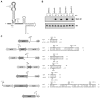
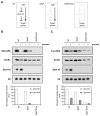
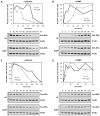
Bacteria selectively consume some carbon sources over others through a regulatory mechanism termed catabolite repression. Here, we show that the base-pairing RNA Spot 42 plays a broad role in catabolite repression in Escherichia coli by directly repressing genes involved in central and secondary metabolism, redox balancing, and the consumption of diverse nonpreferred carbon sources. Many of the genes repressed by Spot 42 are transcriptionally activated by the global regulator CRP. Since CRP represses Spot 42, these regulators participate in a specific regulatory circuit called a multioutput feedforward loop. We found that this loop can reduce leaky expression of target genes in the presence of glucose and can maintain repression of target genes under changing nutrient conditions. Our results suggest that base-pairing RNAs in feedforward loops can help shape the steady-state levels and dynamics of gene expression.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures


Comment in
-
Sweet business: Spot42 RNA networks with CRP to modulate catabolite repression.Mol Cell. 2011 Feb 4;41(3):245-6. doi: 10.1016/j.molcel.2011.01.011. Mol Cell. 2011. PMID: 21292156
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

