Structural basis for dimerization and activity of human PAPD1, a noncanonical poly(A) polymerase
- PMID: 21292163
- PMCID: PMC3057501
- DOI: 10.1016/j.molcel.2011.01.013
Structural basis for dimerization and activity of human PAPD1, a noncanonical poly(A) polymerase
Abstract
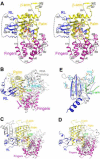
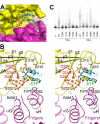
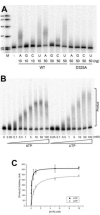
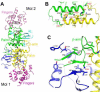
Poly(A) polymerases (PAPs) are found in most living organisms and have important roles in RNA function and metabolism. Here, we report the crystal structure of human PAPD1, a noncanonical PAP that can polyadenylate RNAs in the mitochondria (also known as mtPAP) and oligouridylate histone mRNAs (TUTase1). The overall structure of the palm and fingers domains is similar to that in the canonical PAPs. The active site is located at the interface between the two domains, with a large pocket that can accommodate the substrates. The structure reveals the presence of a previously unrecognized domain in the N-terminal region of PAPD1, with a backbone fold that is similar to that of RNP-type RNA binding domains. This domain (named the RL domain), together with a β-arm insertion in the palm domain, contributes to dimerization of PAPD1. Surprisingly, our mutagenesis and biochemical studies show that dimerization is required for the catalytic activity of PAPD1.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures

References
-
- Adams PD, Grosse-Kunstleve RW, Hung L-W, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building a new software for automated crystallographic structure determination. Acta Cryst. 2002;D58:1948–1954. - PubMed
-
- Balbo PB, Meinke G, Bohm A. Kinetic studies of yeast polyA polymerase indicate an induced fit mechanism for nucleotide specificity. Biochem. 2005;44:7777–7786. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

