Iron-containing transcription factors and their roles as sensors
- PMID: 21292540
- PMCID: PMC3074041
- DOI: 10.1016/j.cbpa.2011.01.006
Iron-containing transcription factors and their roles as sensors
Abstract
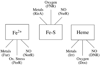
Iron-binding transcription factors are widespread throughout the bacterial world and to date are known to bind several types of cofactors, such as Fe2+, heme, or iron-sulfur clusters. The known chemistry of these cofactors is exploited by transcription factors, including Fur, FNR, and NsrR, to sense molecules such as Fe2+, gases (e.g. oxygen and nitric oxide), or reactive oxygen species. New structural data and information generated by genome-wide analysis studies have provided additional details about the mechanism and function of iron-binding transcription factors that act as sensors.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures
References
-
- Waldron KJ, Robinson NJ. How do bacterial cells ensure that metalloproteins get the correct metal? Nat Rev Microbiol. 2009;7:25–35. - PubMed
-
- Lee JW, Helmann JD. Functional specialization within the Fur family of metalloregulators. Biometals. 2007;20:485–499. - PubMed
-
-
Ahmad R, Brandsdal BO, Michaud-Soret I, Willassen NP. Ferric uptake regulator protein: binding free energy calculations and per-residue free energy decomposition. Proteins: Struct, Funct, and Bioinf. 2009;75:373–386. The authors used computational techniques to determine the site of Fe2+ binding to Fur and the effects of iron ligation. Further, the effects of Fe2+-Fur binding to DNA and the amino acids important for this interaction were investigated.
-
-
- McHugh JP, Rodriguez-Quinones F, Abdul-Tehrani H, Svistunenko DA, Poole RK, Cooper CE, Andrews SC. Global iron-dependent gene regulation in Escherichia coli: a new mechanism for iron homeostasis. J Biol Chem. 2003;278:29478–29486. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

