The low resolution structure of ApoA1 in spherical high density lipoprotein revealed by small angle neutron scattering
- PMID: 21292766
- PMCID: PMC3069452
- DOI: 10.1074/jbc.M110.209130
The low resolution structure of ApoA1 in spherical high density lipoprotein revealed by small angle neutron scattering
Abstract
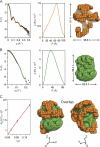
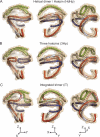
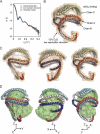
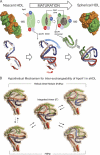
Spherical high density lipoprotein (sHDL), a key player in reverse cholesterol transport and the most abundant form of HDL, is associated with cardiovascular diseases. Small angle neutron scattering with contrast variation was used to determine the solution structure of protein and lipid components of reconstituted sHDL. Apolipoprotein A1, the major protein of sHDL, forms a hollow structure that cradles a central compact lipid core. Three apoA1 chains are arranged within the low resolution structure of the protein component as one of three possible global architectures: (i) a helical dimer with a hairpin (HdHp), (ii) three hairpins (3Hp), or (iii) an integrated trimer (iT) in which the three apoA1 monomers mutually associate over a portion of the sHDL surface. Cross-linking and mass spectrometry analyses help to discriminate among the three molecular models and are most consistent with the HdHp overall architecture of apoA1 within sHDL.
Figures
References
-
- Castelli W. P., Doyle J. T., Gordon T., Hames C. G., Hjortland M. C., Hulley S. B., Kagan A., Zukel W. J. (1977) Circulation 55, 767–772 - PubMed
-
- Durrington P. N., Ishola M., Hunt L., Arrol S., Bhatnagar D. (1988) Lancet 1, 1070–1073 - PubMed
-
- Pekkanen J., Linn S., Heiss G., Suchindran C. M., Leon A., Rifkind B. M., Tyroler H. A. (1990) New Engl. J. Med. 322, 1700–1707 - PubMed
-
- Karathanasis S. K., Norum R. A., Zannis V. I., Breslow J. L. (1983) Nature 301, 718–720 - PubMed
-
- Norum R. A., Lakier J. B., Goldstein S., Angel A., Goldberg R. B., Block W. D., Noffze D. K., Dolphin P. J., Edelglass J., Bogorad D. D., Alaupovic P. (1982) New Engl. J. Med. 306, 1513–1519 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

