Protein native-state stabilization by placing aromatic side chains in N-glycosylated reverse turns
- PMID: 21292975
- PMCID: PMC3099596
- DOI: 10.1126/science.1198461
Protein native-state stabilization by placing aromatic side chains in N-glycosylated reverse turns
Erratum in
- Science. 2012 Mar 2;335(6072):1042
Abstract
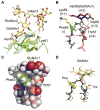
N-glycosylation of eukaryotic proteins helps them fold and traverse the cellular secretory pathway and can increase their stability, although the molecular basis for stabilization is poorly understood. Glycosylation of proteins at naïve sites (ones that normally are not glycosylated) could be useful for therapeutic and research applications but currently results in unpredictable changes to protein stability. We show that placing a phenylalanine residue two or three positions before a glycosylated asparagine in distinct reverse turns facilitates stabilizing interactions between the aromatic side chain and the first N-acetylglucosamine of the glycan. Glycosylating this portable structural module, an enhanced aromatic sequon, in three different proteins stabilizes their native states by -0.7 to -2.0 kilocalories per mole and increases cellular glycosylation efficiency.
Figures

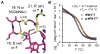
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

