Ribosome recycling depends on a mechanistic link between the FeS cluster domain and a conformational switch of the twin-ATPase ABCE1
- PMID: 21292982
- PMCID: PMC3044390
- DOI: 10.1073/pnas.1015953108
Ribosome recycling depends on a mechanistic link between the FeS cluster domain and a conformational switch of the twin-ATPase ABCE1
Abstract
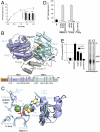
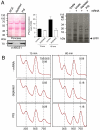
Despite some appealing similarities of protein synthesis across all phyla of life, the final phase of mRNA translation has yet to be captured. Here, we reveal the ancestral role and mechanistic principles of the newly identified twin-ATPase ABCE1 in ribosome recycling. We demonstrate that the unique iron-sulfur cluster domain and an ATP-dependent conformational switch of ABCE1 are essential both for ribosome binding and recycling. By direct (11) interaction, the peptide release factor aRF1 is shown to synergistically promote ABCE1 function in posttermination ribosome recycling. Upon ATP binding, ABCE1 undergoes a conformational switch from an open to a closed ATP-occluded state, which drives ribosome dissociation as well as the disengagement of aRF1. ATP hydrolysis is not required for a single round of ribosome splitting but for ABCE1 release from the 30S subunit to reenter a new cycle. These results provide a mechanistic understanding of final phases in mRNA translation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Dever TE. Gene-specific regulation by general translation factors. Cell. 2002;108:545–556. - PubMed
-
- Ramakrishnan V. Ribosome structure and the mechanism of translation. Cell. 2002;108:557–572. - PubMed
-
- Chakraburtty K. Translational regulation by ABC systems. Res Microbiol. 2001;152:391–399. - PubMed
-
- Schmitt L, Tampé R. Structure and mechanism of ABC transporters. Curr Opin Struct Biol. 2002;12:754–760. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

