Real-time analysis of double-strand DNA break repair by homologous recombination
- PMID: 21292986
- PMCID: PMC3044406
- DOI: 10.1073/pnas.1019660108
Real-time analysis of double-strand DNA break repair by homologous recombination
Abstract
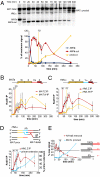
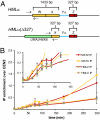
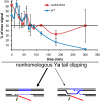
The ability to induce synchronously a single site-specific double-strand break (DSB) in a budding yeast chromosome has made it possible to monitor the kinetics and genetic requirements of many molecular steps during DSB repair. Special attention has been paid to the switching of mating-type genes in Saccharomyces cerevisiae, a process initiated by the HO endonuclease by cleaving the MAT locus. A DSB in MATa is repaired by homologous recombination--specifically, by gene conversion--using a heterochromatic donor, HMLα. Repair results in the replacement of the a-specific sequences (Ya) by Yα and switching from MATa to MATα. We report that MAT switching requires the DNA replication factor Dpb11, although it does not require the Cdc7-Dbf4 kinase or the Mcm and Cdc45 helicase components. Using Southern blot, PCR, and ChIP analysis of samples collected every 10 min, we extend previous studies of this process to identify the times for the loading of Rad51 recombinase protein onto the DSB ends at MAT, the subsequent strand invasion by the Rad51 nucleoprotein filament into the donor sequences, the initiation of new DNA synthesis, and the removal of the nonhomologous Y sequences. In addition we report evidence for the transient displacement of well-positioned nucleosomes in the HML donor locus during strand invasion.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

