Mapping copy number variation by population-scale genome sequencing
- PMID: 21293372
- PMCID: PMC3077050
- DOI: 10.1038/nature09708
Mapping copy number variation by population-scale genome sequencing
Abstract
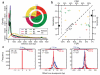
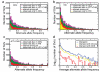
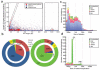
Genomic structural variants (SVs) are abundant in humans, differing from other forms of variation in extent, origin and functional impact. Despite progress in SV characterization, the nucleotide resolution architecture of most SVs remains unknown. We constructed a map of unbalanced SVs (that is, copy number variants) based on whole genome DNA sequencing data from 185 human genomes, integrating evidence from complementary SV discovery approaches with extensive experimental validations. Our map encompassed 22,025 deletions and 6,000 additional SVs, including insertions and tandem duplications. Most SVs (53%) were mapped to nucleotide resolution, which facilitated analysing their origin and functional impact. We examined numerous whole and partial gene deletions with a genotyping approach and observed a depletion of gene disruptions amongst high frequency deletions. Furthermore, we observed differences in the size spectra of SVs originating from distinct formation mechanisms, and constructed a map of SV hotspots formed by common mechanisms. Our analytical framework and SV map serves as a resource for sequencing-based association studies.
Figures


References
Publication types
MeSH terms
Grants and funding
- U54 HG003067/HG/NHGRI NIH HHS/United States
- RC2 HG005552/HG/NHGRI NIH HHS/United States
- G0701805/MRC_/Medical Research Council/United Kingdom
- R01 GM059290/GM/NIGMS NIH HHS/United States
- R01 GM081533/GM/NIGMS NIH HHS/United States
- 077014/WT_/Wellcome Trust/United Kingdom
- 062023/WT_/Wellcome Trust/United Kingdom
- 077009/WT_/Wellcome Trust/United Kingdom
- 077192/WT_/Wellcome Trust/United Kingdom
- R01 HG004719/HG/NHGRI NIH HHS/United States
- G1000758/MRC_/Medical Research Council/United Kingdom
- R01 MH091350/MH/NIMH NIH HHS/United States
- U54 HG003273/HG/NHGRI NIH HHS/United States
- 085532/WT_/Wellcome Trust/United Kingdom
- U01 HG005209/HG/NHGRI NIH HHS/United States
- R21 AA022707/AA/NIAAA NIH HHS/United States
- P41 HG004221/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

