A dynamic model for genome-wide association studies
- PMID: 21293879
- PMCID: PMC3103104
- DOI: 10.1007/s00439-011-0960-6
A dynamic model for genome-wide association studies
Abstract
Although genome-wide association studies (GWAS) are widely used to identify the genetic and environmental etiology of a trait, several key issues related to their statistical power and biological relevance have remained unexplored. Here, we describe a novel statistical approach, called functional GWAS or fGWAS, to analyze the genetic control of traits by integrating biological principles of trait formation into the GWAS framework through mathematical and statistical bridges. fGWAS can address many fundamental questions, such as the patterns of genetic control over development, the duration of genetic effects, as well as what causes developmental trajectories to change or stop changing. In statistics, fGWAS displays increased power for gene detection by capitalizing on cumulative phenotypic variation in a longitudinal trait over time and increased robustness for manipulating sparse longitudinal data.
Figures


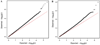
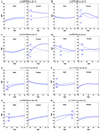

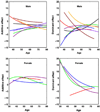
References
-
- Anholt RRH, Mackay TFC. Genetic analysis of complex behaviors in Drosophila. Nat Rev Genet. 2004;5:838–849. - PubMed
-
- Bock RD, Thissen D. Fitting multi-component models for growth in stature; Proceedings of the 9th international biometrics conference; 1976. pp. 431–442.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

