Noninvasive genomic detection of melanoma
- PMID: 21294715
- PMCID: PMC3118279
- DOI: 10.1111/j.1365-2133.2011.10239.x
Noninvasive genomic detection of melanoma
Abstract
Background: Early detection and treatment of melanoma is important for optimal clinical outcome, leading to biopsy of pigmented lesions deemed suspicious for the disease. The vast majority of such lesions are benign. Thus, a more objective and accurate means for detection of melanoma is needed to identify lesions for excision.
Objectives: To provide proof-of-principle that epidermal genetic information retrieval (EGIR™; DermTech International, La Jolla, CA, U.S.A.), a method that noninvasively samples cells from stratum corneum by means of adhesive tape stripping, can be used to discern melanomas from naevi.
Methods: Skin overlying pigmented lesions clinically suspicious for melanoma was harvested using EGIR. RNA isolated from the tapes was amplified and gene expression profiled. All lesions were removed for histopathological evaluation.
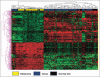
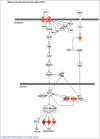
Results: Supervised analysis of the microarray data identified 312 genes differentially expressed between melanomas, naevi and normal skin specimens (P<0·001, false discovery rate q<0·05). Surprisingly, many of these genes are known to have a role in melanocyte development and physiology, melanoma, cancer, and cell growth control. Subsequent class prediction modelling of a training dataset, consisting of 37 melanomas and 37 naevi, discovered a 17-gene classifier that discriminates these skin lesions. Upon testing with an independent dataset, this classifier discerned in situ and invasive melanomas from naevi with 100% sensitivity and 88% specificity, with an area under the curve for the receiver operating characteristic of 0·955.
Conclusions: These results demonstrate that EGIR-harvested specimens can be used to detect melanoma accurately by means of a 17-gene genomic biomarker.
© 2011 The Authors. BJD © 2011 British Association of Dermatologists 2011.
Figures

Comment in
-
Stratum corneum RNA levels are diagnostic for melanoma.Br J Dermatol. 2011 Apr;164(4):693-4. doi: 10.1111/j.1365-2133.2011.10283.x. Br J Dermatol. 2011. PMID: 21457197 No abstract available.
-
Paratumoral gene expression profiles: promising markers of malignancy in melanocytic lesions.Br J Dermatol. 2011 Sep;165(3):702-3. doi: 10.1111/j.1365-2133.2011.10437.x. Epub 2011 Jul 22. Br J Dermatol. 2011. PMID: 21623754 No abstract available.
References
-
- Lens MB, Dawes M. Global perspectives of contemporary epidemiological trends of cutaneous malignant melanoma. Br J Dermatol. 2004;150:179–85. - PubMed
-
- Altekruse SF, Kosary CL, Krapcho M, et al. SEER Cancer Statistics Review, 1975–2007. Bethesda, MD: National Cancer Institute; 2010.
-
- Minor DR, Moore D, Kim C, et al. Prognostic factors in metastatic melanoma patients treated with biochemotherapy and maintenance immunotherapy. Oncologist. 2009;14:995–1002. - PubMed
-
- Goldsmith LA, Askin FB, Chang AE, et al. Diagnosis and treatment of early melanoma: NIH Consensus Development Panel on Early Melanoma. JAMA. 1992;268:1314–19.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

