Independent component and pathway-based analysis of miRNA-regulated gene expression in a model of type 1 diabetes
- PMID: 21294859
- PMCID: PMC3040732
- DOI: 10.1186/1471-2164-12-97
Independent component and pathway-based analysis of miRNA-regulated gene expression in a model of type 1 diabetes
Abstract
Background: Several approaches have been developed for miRNA target prediction, including methods that incorporate expression profiling. However the methods are still in need of improvements due to a high false discovery rate. So far, none of the methods have used independent component analysis (ICA). Here, we developed a novel target prediction method based on ICA that incorporates both seed matching and expression profiling of miRNA and mRNA expressions. The method was applied on a cellular model of type 1 diabetes.
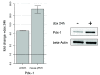
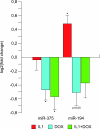
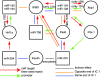
Results: Microarray profiling identified eight miRNAs (miR-124/128/192/194/204/375/672/708) with differential expression. Applying ICA on the mRNA profiling data revealed five significant independent components (ICs) correlating to the experimental conditions. The five ICs also captured the miRNA expressions by explaining > 97% of their variance. By using ICA, seven of the eight miRNAs showed significant enrichment of sequence predicted targets, compared to only four miRNAs when using simple negative correlation. The ICs were enriched for miRNA targets that function in diabetes-relevant pathways e.g. type 1 and type 2 diabetes and maturity onset diabetes of the young (MODY).
Conclusions: In this study, ICA was applied as an attempt to separate the various factors that influence the mRNA expression in order to identify miRNA targets. The results suggest that ICA is better at identifying miRNA targets than negative correlation. Additionally, combining ICA and pathway analysis constitutes a means for prioritizing between the predicted miRNA targets. Applying the method on a model of type 1 diabetes resulted in identification of eight miRNAs that appear to affect pathways of relevance to disease mechanisms in diabetes.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

