RB1, development, and cancer
- PMID: 21295686
- PMCID: PMC3691055
- DOI: 10.1016/B978-0-12-380916-2.00005-X
RB1, development, and cancer
Abstract
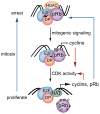
The RB1 gene is the first tumor suppressor gene identified whose mutational inactivation is the cause of a human cancer, the pediatric cancer retinoblastoma. The 25 years of research since its discovery has not only illuminated a general role for RB1 in human cancer, but also its critical importance in normal development. Understanding the molecular function of the RB1 encoded protein, pRb, is a long-standing goal that promises to inform our understanding of cancer, its relationship to normal development, and possible therapeutic strategies to combat this disease. Achieving this goal has been difficult, complicated by the complexity of pRb and related proteins. The goal of this review is to explore the hypothesis that, at its core, the molecular function of pRb is to dynamically regulate the location-specific assembly or disassembly of protein complexes on the DNA in response to the output of various signaling pathways. These protein complexes participate in a variety of molecular processes relevant to DNA including gene transcription, DNA replication, DNA repair, and mitosis. Through regulation of these processes, RB1 plays a uniquely prominent role in normal development and cancer.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures


References
-
- Aggarwal BD, Calvi BR. Chromatin regulates origin activity in Drosophila follicle cells. Nature. 2004;430:372–6. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

