Influence of cell geometry on division-plane positioning
- PMID: 21295701
- PMCID: PMC3048034
- DOI: 10.1016/j.cell.2011.01.016
Influence of cell geometry on division-plane positioning
Abstract
The spatial organization of cells depends on their ability to sense their own shape and size. Here, we investigate how cell shape affects the positioning of the nucleus, spindle and subsequent cell division plane. To manipulate geometrical parameters in a systematic manner, we place individual sea urchin eggs into microfabricated chambers of defined geometry (e.g., triangles, rectangles, and ellipses). In each shape, the nucleus is positioned at the center of mass and is stretched by microtubules along an axis maintained through mitosis and predictive of the future division plane. We develop a simple computational model that posits that microtubules sense cell geometry by probing cellular space and orient the nucleus by exerting pulling forces that scale to microtubule length. This model quantitatively predicts division-axis orientation probability for a wide variety of cell shapes, even in multicellular contexts, and estimates scaling exponents for length-dependent microtubule forces.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures
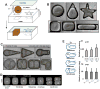

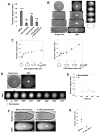



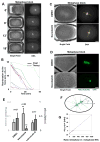
Comment in
-
Getting cells and tissues into shape.Cell. 2011 Feb 4;144(3):325-6. doi: 10.1016/j.cell.2011.01.022. Cell. 2011. PMID: 21295695
References
-
- Bettinger BT, Gilbert DM, Amberg DC. Actin up in the nucleus. Nat Rev Mol Cell Biol. 2004;5:410–415. - PubMed
-
- Bjerknes M. Physical theory of the orientation of astral mitotic spindles. Science. 1986;234:1413–1416. - PubMed
-
- Concha ML, Adams RJ. Oriented cell divisions and cellular morphogenesis in the zebrafish gastrula and neurula: a time-lapse analysis. Development. 1998;125:983–994. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

