A TACC3/ch-TOG/clathrin complex stabilises kinetochore fibres by inter-microtubule bridging
- PMID: 21297582
- PMCID: PMC3049211
- DOI: 10.1038/emboj.2011.15
A TACC3/ch-TOG/clathrin complex stabilises kinetochore fibres by inter-microtubule bridging
Abstract
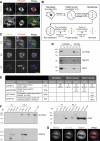
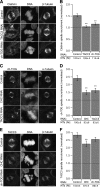
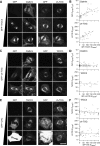
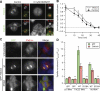
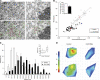
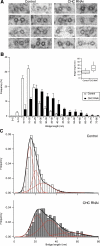
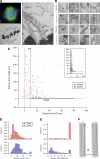
Kinetochore fibres (K-fibres) of the spindle apparatus move chromosomes during mitosis. These fibres are discrete bundles of parallel microtubules (MTs) that are crosslinked by inter-MT 'bridges' that are thought to improve fibre stability during chromosomal movement. The identity of these bridges is unknown. Clathrin is a multimeric protein that has been shown to stabilise K-fibres during early mitosis by a mechanism independent of its role in membrane trafficking. In this study, we show that clathrin at the mitotic spindle is in a transforming acidic colied-coil protein 3 (TACC3)/colonic, hepatic tumour overexpressed gene (ch-TOG)/clathrin complex. The complex is anchored to the spindle by TACC3 and ch-TOG. Ultrastructural analysis of clathrin-depleted K-fibres revealed a selective loss of a population of short inter-MT bridges and a general loss of MTs. A similar loss of short inter-MT bridges was observed in TACC3-depleted K-fibres. Finally, immunogold labelling confirmed that inter-MT bridges in K-fibres contain clathrin. Our results suggest that the TACC3/ch-TOG/clathrin complex is an inter-MT bridge that stabilises K-fibres by physical crosslinking and by reducing rates of MT catastrophe.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
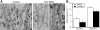

References
-
- Barr AR, Gergely F (2007) Aurora-A: the maker and breaker of spindle poles. J Cell Sci 120: 2987–2996 - PubMed
-
- Bieling P, Telley IA, Surrey T (2010) A minimal midzone protein module controls formation and length of antiparallel microtubule overlaps. Cell 142: 420–432 - PubMed
-
- Brodsky FM, Chen CY, Knuehl C, Towler MC, Wakeham DE (2001) Biological basket weaving: formation and function of clathrin-coated vesicles. Annu Rev Cell Dev Biol 17: 517–568 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

