Genome-wide mapping of Arabidopsis thaliana origins of DNA replication and their associated epigenetic marks
- PMID: 21297636
- PMCID: PMC3079358
- DOI: 10.1038/nsmb.1988
Genome-wide mapping of Arabidopsis thaliana origins of DNA replication and their associated epigenetic marks
Abstract
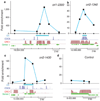
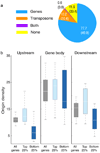
Genome integrity requires faithful chromosome duplication. Origins of replication, the genomic sites at which DNA replication initiates, are scattered throughout the genome. Their mapping at a genomic scale in multicellular organisms has been challenging. In this study we profiled origins in Arabidopsis thaliana by high-throughput sequencing of newly synthesized DNA and identified ~1,500 putative origins genome-wide. This was supported by chromatin immunoprecipitation and microarray (ChIP-chip) experiments to identify ORC1- and CDC6-binding sites. We validated origin activity independently by measuring the abundance of nascent DNA strands. The midpoints of most A. thaliana origin regions are preferentially located within the 5' half of genes, enriched in G+C, histone H2A.Z, H3K4me2, H3K4me3 and H4K5ac, and depleted in H3K4me1 and H3K9me2. Our data help clarify the epigenetic specification of DNA replication origins in A. thaliana and have implications for other eukaryotes.
Figures



References
-
- Huberman JA, Riggs AD. On the mechanism of DNA replication in mammalian chromosomes. J Mol Biol. 1968;32:327–341. - PubMed
-
- DePamphilis ML, et al. Regulating the licensing of DNA replication origins in metazoa. Curr. Opin. Cell Biol. 2006;18:231–239. - PubMed
-
- Aladjem MI. Replication in context: dynamic regulation of DNA replication patterns in metazoans. Nat Rev Genet. 2007;8:588–600. - PubMed
-
- Bell SP, Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992;357:128–134. - PubMed
-
- Wyrick JJ, et al. Genome-wide distribution of ORC and MCM proteins in S. cerevisiae: high-resolution mapping of replication origins. Science. 2001;294:2357–2360. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

