Comparative and joint analysis of two metagenomic datasets from a biogas fermenter obtained by 454-pyrosequencing
- PMID: 21297863
- PMCID: PMC3027613
- DOI: 10.1371/journal.pone.0014519
Comparative and joint analysis of two metagenomic datasets from a biogas fermenter obtained by 454-pyrosequencing
Abstract
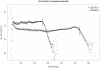
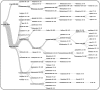
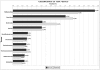
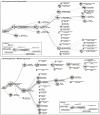
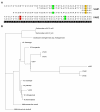
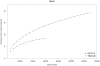
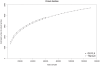
Biogas production from renewable resources is attracting increased attention as an alternative energy source due to the limited availability of traditional fossil fuels. Many countries are promoting the use of alternative energy sources for sustainable energy production. In this study, a metagenome from a production-scale biogas fermenter was analysed employing Roche's GS FLX Titanium technology and compared to a previous dataset obtained from the same community DNA sample that was sequenced on the GS FLX platform. Taxonomic profiling based on 16S rRNA-specific sequences and an Environmental Gene Tag (EGT) analysis employing CARMA demonstrated that both approaches benefit from the longer read lengths obtained on the Titanium platform. Results confirmed Clostridia as the most prevalent taxonomic class, whereas species of the order Methanomicrobiales are dominant among methanogenic Archaea. However, the analyses also identified additional taxa that were missed by the previous study, including members of the genera Streptococcus, Acetivibrio, Garciella, Tissierella, and Gelria, which might also play a role in the fermentation process leading to the formation of methane. Taking advantage of the CARMA feature to correlate taxonomic information of sequences with their assigned functions, it appeared that Firmicutes, followed by Bacteroidetes and Proteobacteria, dominate within the functional context of polysaccharide degradation whereas Methanomicrobiales represent the most abundant taxonomic group responsible for methane production. Clostridia is the most important class involved in the reductive CoA pathway (Wood-Ljungdahl pathway) that is characteristic for acetogenesis. Based on binning of 16S rRNA-specific sequences allocated to the dominant genus Methanoculleus, it could be shown that this genus is represented by several different species. Phylogenetic analysis of these sequences placed them in close proximity to the hydrogenotrophic methanogen Methanoculleus bourgensis. While rarefaction analyses still indicate incomplete coverage, examination of the GS FLX Titanium dataset resulted in the identification of additional genera and functional elements, providing a far more complete coverage of the community involved in anaerobic fermentative pathways leading to methane formation.
Conflict of interest statement
Figures

Similar articles
-
Phylogenetic characterization of a biogas plant microbial community integrating clone library 16S-rDNA sequences and metagenome sequence data obtained by 454-pyrosequencing.J Biotechnol. 2009 Jun 1;142(1):38-49. doi: 10.1016/j.jbiotec.2009.02.010. Epub 2009 Feb 27. J Biotechnol. 2009. PMID: 19480946
-
Comparative metagenomics of biogas-producing microbial communities from production-scale biogas plants operating under wet or dry fermentation conditions.Biotechnol Biofuels. 2015 Feb 8;8:14. doi: 10.1186/s13068-014-0193-8. eCollection 2015. Biotechnol Biofuels. 2015. PMID: 25688290 Free PMC article.
-
The metagenome of a biogas-producing microbial community of a production-scale biogas plant fermenter analysed by the 454-pyrosequencing technology.J Biotechnol. 2008 Aug 31;136(1-2):77-90. doi: 10.1016/j.jbiotec.2008.05.008. Epub 2008 May 27. J Biotechnol. 2008. PMID: 18597880
-
Metagenome, metatranscriptome, and metaproteome approaches unraveled compositions and functional relationships of microbial communities residing in biogas plants.Appl Microbiol Biotechnol. 2018 Jun;102(12):5045-5063. doi: 10.1007/s00253-018-8976-7. Epub 2018 Apr 30. Appl Microbiol Biotechnol. 2018. PMID: 29713790 Free PMC article. Review.
-
Metaproteomics of complex microbial communities in biogas plants.Microb Biotechnol. 2015 Sep;8(5):749-63. doi: 10.1111/1751-7915.12276. Epub 2015 Apr 15. Microb Biotechnol. 2015. PMID: 25874383 Free PMC article. Review.
Cited by
-
Noteworthy Facts about a Methane-Producing Microbial Community Processing Acidic Effluent from Sugar Beet Molasses Fermentation.PLoS One. 2015 May 22;10(5):e0128008. doi: 10.1371/journal.pone.0128008. eCollection 2015. PLoS One. 2015. PMID: 26000448 Free PMC article.
-
Functional and Structural Succession of Soil Microbial Communities below Decomposing Human Cadavers.PLoS One. 2015 Jun 12;10(6):e0130201. doi: 10.1371/journal.pone.0130201. eCollection 2015. PLoS One. 2015. PMID: 26067226 Free PMC article.
-
Temporal variation in bacterial and methanogenic communities of three full-scale anaerobic digesters treating swine wastewater.Environ Sci Pollut Res Int. 2019 Jan;26(2):1217-1226. doi: 10.1007/s11356-017-1103-y. Epub 2018 Jan 11. Environ Sci Pollut Res Int. 2019. PMID: 29327188
-
Metagenome from a Spirulina digesting biogas reactor: analysis via binning of contigs and classification of short reads.BMC Microbiol. 2015 Dec 17;15:277. doi: 10.1186/s12866-015-0615-1. BMC Microbiol. 2015. PMID: 26680455 Free PMC article.
-
Characterization of a biogas-producing microbial community by short-read next generation DNA sequencing.Biotechnol Biofuels. 2012 Jul 12;5:41. doi: 10.1186/1754-6834-5-41. eCollection 2012. Biotechnol Biofuels. 2012. PMID: 22673110 Free PMC article.
References
-
- Weiland P. Production and energetic use of biogas from energy crops and wastes in Germany. Applied Biochemistry and Biotechnology. 2003;109:263–274. - PubMed
-
- Yadvika S, Sreekrishnan T, Kohli S, Rana V. Enhancement of biogas production from solid substrates using different techniques - a review. Bioresource Technology. 2004;95:1–10. - PubMed
-
- Bayer E, Belaich J, Shoham Y, Lamed R. The cellulosomes: multienzyme machines for degradation of plant cell wall polysaccharides. Annual Review of Microbiology. 2004;58:521–54. - PubMed
-
- Cirne D, Lehtomaki A, Bjornsson L, Blackall L. Hydrolysis and microbial community analyses in two-stage anaerobic digestion of energy crops. Journal of Applied Microbiology. 2007;103:516–527. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

