Molecular evolutionary analysis of ABCB5: the ancestral gene is a full transporter with potentially deleterious single nucleotide polymorphisms
- PMID: 21298007
- PMCID: PMC3029322
- DOI: 10.1371/journal.pone.0016318
Molecular evolutionary analysis of ABCB5: the ancestral gene is a full transporter with potentially deleterious single nucleotide polymorphisms
Abstract
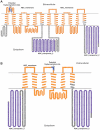
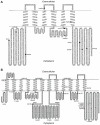
Background: ABCB5 is a member of the ABC protein superfamily, which includes the transporters ABCB1, ABCC1 and ABCG2 responsible for causing drug resistance in cancer patients and also several other transporters that have been linked to human disease. The ABCB5 full transporter (ABCB5.ts) is expressed in human testis and its functional significance is presently unknown. Another variant of this transporter, ABCB5 beta possess a "half-transporter-like" structure and is expressed in melanoma stem cells, normal melanocytes, and other types of pigment cells. ABCB5 beta has important clinical implications, as it may be involved with multidrug resistance in melanoma stem cells, allowing these stem cells to survive chemotherapeutic regimes.
Methodology/principal findings: We constructed and examined in detail topological structures of the human ABCB5 protein and determined in-silico the cSNPs (coding single nucleotide polymorphisms) that may affect its function. Evolutionary analysis of ABCB5 indicated that ABCB5, ABCB1, ABCB4, and ABCB11 share a common ancestor, which began duplicating early in the evolutionary history of chordates. This suggests that ABCB5 has evolved as a full transporter throughout its evolutionary history.
Conclusions/significance: From our in-silco analysis of cSNPs we found that a large number of non-synonymous cSNPs map to important functional regions of the protein suggesting that these SNPs if present in human populations may play a role in diseases associated with ABCB5. From phylogenetic analyses, we have shown that ABCB5 evolved as a full transporter throughout its evolutionary history with an absence of any major shifts in selection between the various lineages suggesting that the function of ABCB5 has been maintained during mammalian evolution. This finding would suggest that ABCB5 beta may have evolved to play a specific role in human pigment cells and/or melanoma cells where it is predominantly expressed.
Conflict of interest statement
Figures




References
-
- Dean M. ABC transporters, drug resistance, and cancer stem cells. J Mammary Gland Biol Neoplasia. 2009;14:3–9. - PubMed
-
- Calcagno AM, Kim IW, Wu CP, Shukla S, Ambudkar SV. ABC drug transporters as molecular targets for the prevention of multidrug resistance and drug–drug interactions. Curr Drug Deliv. 2007;4:324333. - PubMed
-
- Gottesman MM, Fojo T, Bates SE. Multidrug resistance in cancer: role of ATP–dependent transporters. Nat Rev Cancer. 2002;2:48–58. - PubMed
-
- Dean M, Rzhetsky A, Allikmets R. The human ATP-binding cassette (ABC) transporter superfamily. Genome Res. 2001;11:1156–1166. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

