Horizontal transfer, not duplication, drives the expansion of protein families in prokaryotes
- PMID: 21298028
- PMCID: PMC3029252
- DOI: 10.1371/journal.pgen.1001284
Horizontal transfer, not duplication, drives the expansion of protein families in prokaryotes
Abstract
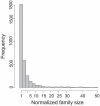
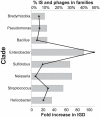
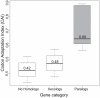
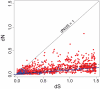
Gene duplication followed by neo- or sub-functionalization deeply impacts the evolution of protein families and is regarded as the main source of adaptive functional novelty in eukaryotes. While there is ample evidence of adaptive gene duplication in prokaryotes, it is not clear whether duplication outweighs the contribution of horizontal gene transfer in the expansion of protein families. We analyzed closely related prokaryote strains or species with small genomes (Helicobacter, Neisseria, Streptococcus, Sulfolobus), average-sized genomes (Bacillus, Enterobacteriaceae), and large genomes (Pseudomonas, Bradyrhizobiaceae) to untangle the effects of duplication and horizontal transfer. After removing the effects of transposable elements and phages, we show that the vast majority of expansions of protein families are due to transfer, even among large genomes. Transferred genes--xenologs--persist longer in prokaryotic lineages possibly due to a higher/longer adaptive role. On the other hand, duplicated genes--paralogs--are expressed more, and, when persistent, they evolve slower. This suggests that gene transfer and gene duplication have very different roles in shaping the evolution of biological systems: transfer allows the acquisition of new functions and duplication leads to higher gene dosage. Accordingly, we show that paralogs share most protein-protein interactions and genetic regulators, whereas xenologs share very few of them. Prokaryotes invented most of life's biochemical diversity. Therefore, the study of the evolution of biology systems should explicitly account for the predominant role of horizontal gene transfer in the diversification of protein families.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- McCutcheon JP, McDonald BR, Moran NA. Origin of an alternative genetic code in the extremely small and GC-rich genome of a bacterial symbiont. PLoS Genet. 2009;5:e1000565. doi: 10.1371/journal.pgen.1000565. - DOI - PMC - PubMed
-
- Schneiker S, Perlova O, Kaiser O, Gerth K, Alici A, et al. Complete genome sequence of the myxobacterium Sorangium cellulosum. Nat Biotechnol. 2007;25:1281–1289. - PubMed
-
- Pasek S, Risler JL, Brezellec P. The role of domain redundancy in genetic robustness against null mutations. J Mol Biol. 2006;362:184–191. - PubMed
-
- Wagner A. Gene duplications, robustness and evolutionary innovations. Bioessays. 2008;30:367–373. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

