Evolutionary conserved regulation of HIF-1β by NF-κB
- PMID: 21298084
- PMCID: PMC3029248
- DOI: 10.1371/journal.pgen.1001285
Evolutionary conserved regulation of HIF-1β by NF-κB
Abstract
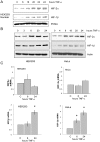
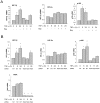
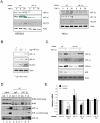
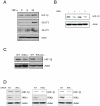
Hypoxia Inducible Factor-1 (HIF-1) is essential for mammalian development and is the principal transcription factor activated by low oxygen tensions. HIF-α subunit quantities and their associated activity are regulated in a post-translational manner, through the concerted action of a class of enzymes called Prolyl Hydroxylases (PHDs) and Factor Inhibiting HIF (FIH) respectively. However, alternative modes of HIF-α regulation such as translation or transcription are under-investigated, and their importance has not been firmly established. Here, we demonstrate that NF-κB regulates the HIF pathway in a significant and evolutionary conserved manner. We demonstrate that NF-κB directly regulates HIF-1β mRNA and protein. In addition, we found that NF-κB-mediated changes in HIF-1β result in modulation of HIF-2α protein. HIF-1β overexpression can rescue HIF-2α protein levels following NF-κB depletion. Significantly, NF-κB regulates HIF-1β (tango) and HIF-α (sima) levels and activity (Hph/fatiga, ImpL3/ldha) in Drosophila, both in normoxia and hypoxia, indicating an evolutionary conserved mode of regulation. These results reveal a novel mechanism of HIF regulation, with impact in the development of novel therapeutic strategies for HIF-related pathologies including ageing, ischemia, and cancer.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Rocha S. Gene regulation under low oxygen: holding your breath for transcription. Trends Biochem Sci. 2007;32:389–397. - PubMed
-
- Kenneth NS, Rocha S. Regulation of gene expression by hypoxia. Biochem J. 2008;414:19–29. - PubMed
-
- Fandrey J, Gorr TA, Gassmann M. Regulating cellular oxygen sensing by hydroxylation. Cardiovasc Res. 2006;71:642–651. - PubMed
-
- Gorr TA, Gassmann M, Wappner P. Sensing and responding to hypoxia via HIF in model invertebrates. J Insect Physiol. 2006;52:349–364. - PubMed
-
- Epstein AC, Gleadle JM, McNeill LA, Hewitson KS, O'Rourke J, et al. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell. 2001;107:43–54. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

