Function of the usher N-terminus in catalysing pilus assembly
- PMID: 21299650
- PMCID: PMC3079318
- DOI: 10.1111/j.1365-2958.2010.07505.x
Function of the usher N-terminus in catalysing pilus assembly
Abstract
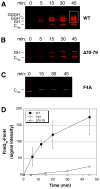
The chaperone/usher (CU) pathway is a conserved bacterial secretion system that assembles adhesive fibres termed pili or fimbriae. Pilus biogenesis by the CU pathway requires a periplasmic chaperone and an outer membrane (OM) assembly platform termed the usher. The usher catalyses formation of subunit-subunit interactions to promote polymerization of the pilus fibre and provides the channel for fibre secretion. The mechanism by which the usher catalyses pilus assembly is not known. Using the P and type 1 pilus systems of uropathogenic Escherichia coli, we show that a conserved N-terminal disulphide region of the PapC and FimD ushers, as well as residue F4 of FimD, are required for the catalytic activity of the ushers. PapC disulphide loop mutants were able to bind PapDG chaperone-subunit complexes, but did not assemble PapG into pilus fibres. FimD disulphide loop and F4 mutants were able to bind chaperone-subunit complexes and initiate assembly of pilus fibres, but were defective for extending the pilus fibres, as measured using in vivo co-purification and in vitro pilus polymerization assays. These results suggest that the catalytic activity of PapC is required to initiate pilus biogenesis, whereas the catalytic activity of FimD is required for extension of the pilus fibre.
© 2010 Blackwell Publishing Ltd.
Figures



References
-
- Baga M, Norgren M, Normark S. Biogenesis of E. coli Pap pili: PapH, a minor pilin subunit involved in cell anchoring and length modulation. Cell. 1987;49:241–251. - PubMed
-
- Blomfield IC, McClain MS, Eisenstein BI. Type 1 fimbriae mutants of Escherichia coli K12: characterization of recognized afimbriate strains and construction of new fim deletion mutants. Mol Microbiol. 1991;5:1439–1445. - PubMed
-
- Choudhury D, Thompson A, Stojanoff V, Langermann S, Pinkner J, Hultgren SJ, Knight SD. X-ray structure of the FimC-FimH chaperone-adhesin complex from uropathogenic Escherichia coli. Science. 1999;285:1061–1066. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

