Molecular mimicry and ligand recognition in binding and catalysis by the histone demethylase LSD1-CoREST complex
- PMID: 21300290
- PMCID: PMC3059804
- DOI: 10.1016/j.str.2011.01.001
Molecular mimicry and ligand recognition in binding and catalysis by the histone demethylase LSD1-CoREST complex
Abstract
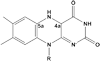
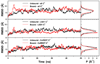
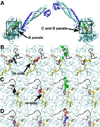
Histone demethylases LSD1 and LSD2 (KDM1A/B) catalyze the oxidative demethylation of Lys4 of histone H3. We used molecular dynamics simulations to probe the diffusion of the oxygen substrate. Oxygen can reach the catalytic center independently from the presence of a bound histone peptide, implying that LSD1 can complete subsequent demethylation cycles without detaching from the nucleosomal particle. The simulations highlight the role of a strictly conserved active-site Lys residue providing general insight into the enzymatic mechanism of oxygen-reacting flavoenzymes. The crystal structure of LSD1-CoREST bound to a peptide of the transcription factor SNAIL1 unravels a fascinating example of molecular mimicry. The SNAIL1 N-terminal residues bind to the enzyme active-site cleft, effectively mimicking the H3 tail. This finding predicts that other members of the SNAIL/Scratch transcription factor family might associate to LSD1/2. The combination of selective histone-modifying activity with the distinct recognition mechanisms underlies the biological complexity of LSD1/2.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures



References
-
- Amente S, Bertoni A, Morano A, Lania L, Avvedimento EV, Majello B. LSD1-mediated demethylation of histone H3 lysine 4 triggers Myc-induced transcription. Oncogene. 2010;29:3691–3702. - PubMed
-
- Åqvist J. Ion Water Interaction Potentials Derived from Free-Energy Perturbation Simulations. J. Phys. Chem. 1990;94:8021–8024.
-
- Baron R, McCammon JA, Mattevi A. The oxygen-binding vs. oxygen-consuming paradigm in biocatalysis: structural biology and biomolecular simulation. Curr. Opin. Struct. Biol. 2009a;19:672–679. - PubMed
-
- Baron R, Trzesniak D, de Vries AH, Elsener A, Marrink SJ, van Gunsteren WF. Comparison of thermodynamic properties of coarse-grained and atomic-level simulation models. ChemPhysChem. 2007;8:452–461. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

