Structure of the putative dihydroorotate dehydrogenase from Streptococcus mutans
- PMID: 21301083
- PMCID: PMC3034605
- DOI: 10.1107/S1744309110048414
Structure of the putative dihydroorotate dehydrogenase from Streptococcus mutans
Abstract
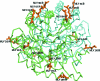
Streptococcus mutans is one of the pathogenic species involved in dental caries, especially in the initiation and development stages. Here, the crystal structure of SMU.595, a putative dihydroorotate dehydrogenase (DHOD) from S. mutans, is reported at 2.4 Å resolution. DHOD is a flavin mononucleotide-containing enzyme which catalyzes the oxidation of L-dihydroorotate to orotate, which is the fourth step and the only redox reaction in the de novo biosynthesis of pyrimidine nucleotides. The reductive lysine-methylation procedure was applied in order to improve the diffraction qualities of the crystals. Analysis of the S. mutans DHOD crystal structure shows that this enzyme is a class 1A DHOD and also suggests potential sites that could be exploited for the design of highly specific inhibitors using the structure-based chemotherapeutic design technique.
Figures



Similar articles
-
Expression, purification and crystallization of Trypanosoma cruzi dihydroorotate dehydrogenase complexed with orotate.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2005 Oct 1;61(Pt 10):875-8. doi: 10.1107/S174430910502659X. Epub 2005 Sep 13. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2005. PMID: 16511183 Free PMC article.
-
Inhibitor binding in a class 2 dihydroorotate dehydrogenase causes variations in the membrane-associated N-terminal domain.Protein Sci. 2004 Apr;13(4):1031-42. doi: 10.1110/ps.03533004. Protein Sci. 2004. PMID: 15044733 Free PMC article.
-
Structures of Trypanosoma cruzi dihydroorotate dehydrogenase complexed with substrates and products: atomic resolution insights into mechanisms of dihydroorotate oxidation and fumarate reduction.Biochemistry. 2008 Oct 14;47(41):10881-91. doi: 10.1021/bi800413r. Epub 2008 Sep 23. Biochemistry. 2008. PMID: 18808149
-
[Structural characteristics and catalytic cycle of dihydroorotate dehydrogenase-a review].Sheng Wu Gong Cheng Xue Bao. 2020 Dec 25;36(12):2732-2740. doi: 10.13345/j.cjb.200166. Sheng Wu Gong Cheng Xue Bao. 2020. PMID: 33398968 Review. Chinese.
-
Target sites for the design of anti-trypanosomatid drugs based on the structure of dihydroorotate dehydrogenase.Curr Pharm Des. 2013;19(14):2615-27. doi: 10.2174/1381612811319140011. Curr Pharm Des. 2013. PMID: 23116399 Review.
Cited by
-
Characterisation of putative class 1A DHODH-like proteins from Mucorales and dematiaceous mould species.PLoS One. 2023 Aug 2;18(8):e0289441. doi: 10.1371/journal.pone.0289441. eCollection 2023. PLoS One. 2023. PMID: 37531380 Free PMC article.
-
Structural and Biochemical Features of Eimeria tenella Dihydroorotate Dehydrogenase, a Potential Drug Target.Genes (Basel). 2020 Dec 7;11(12):1468. doi: 10.3390/genes11121468. Genes (Basel). 2020. PMID: 33297567 Free PMC article.
References
-
- Bradford, M. M. (1976). Anal. Biochem. 72, 248–254. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources

