Structures of β-hairpin antimicrobial protegrin peptides in lipopolysaccharide membranes: mechanism of gram selectivity obtained from solid-state nuclear magnetic resonance
- PMID: 21302955
- PMCID: PMC3062705
- DOI: 10.1021/bi101975v
Structures of β-hairpin antimicrobial protegrin peptides in lipopolysaccharide membranes: mechanism of gram selectivity obtained from solid-state nuclear magnetic resonance
Abstract
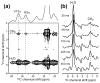
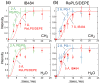
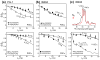
The structural basis for the gram selectivity of two disulfide-bonded β-hairpin antimicrobial peptides (AMPs) is investigated using solid-state nuclear magnetic resonance (NMR) spectroscopy. The hexa-arginine PG-1 exhibits potent activities against both gram-positive and gram-negative bacteria, while a mutant of PG-1 with only three cationic residues maintains gram-positive activity but is 30-fold less active against gram-negative bacteria. We determined the topological structure and lipid interactions of these two peptides in a lipopolysaccharide (LPS)-rich membrane that mimics the outer membrane of gram-negative bacteria and in the POPE/POPG membrane, which mimics the membrane of gram-positive bacteria. (31)P NMR line shapes indicate that both peptides cause less orientational disorder in the LPS-rich membrane than in the POPE/POPG membrane. (13)C chemical shifts and (13)C-(1)H dipolar couplings show that both peptides maintain their β-hairpin conformation in these membranes and are largely immobilized, but the mutant exhibits noticeable intermediate-time scale motion in the LPS membrane at physiological temperature, suggesting shallow insertion. Indeed, (1)H spin diffusion from lipid chains to the peptides shows that PG-1 fully inserts into the LPS-rich membrane whereas the mutant does not. The (13)C-(31)P distances between the most hydrophobically embedded Arg of PG-1 and the lipid (31)P are significantly longer in the LPS membrane than in the POPE/POPG membrane, indicating that PG-1 does not cause toroidal pore defects in the LPS membrane, in contrast to its behavior in the POPE/POPG membrane. Taken together, these data indicate that PG-1 causes transmembrane pores of the barrel-stave type in the LPS membrane, thus allowing further translocation of the peptide into the inner membrane of gram-negative bacteria to kill the cells. In comparison, the less cationic mutant cannot fully cross the LPS membrane because of weaker electrostatic attractions, thus causing weaker antimicrobial activities. Therefore, strong electrostatic attraction between the peptide and the membrane surface, ensured by having a sufficient number of Arg residues, is essential for potent antimicrobial activities against gram-negative bacteria. The data provide a rational basis for controlling gram selectivity of AMPs by adjusting the charge densities.
Figures







Similar articles
-
A 2H solid-state NMR study of lipid clustering by cationic antimicrobial and cell-penetrating peptides in model bacterial membranes.Biophys J. 2013 Nov 19;105(10):2333-42. doi: 10.1016/j.bpj.2013.08.020. Biophys J. 2013. PMID: 24268145 Free PMC article.
-
Antimicrobial peptide interactions with bacterial cell membranes.J Biomol Struct Dyn. 2025 Jun;43(9):4615-4628. doi: 10.1080/07391102.2024.2304683. Epub 2024 Jan 23. J Biomol Struct Dyn. 2025. PMID: 38263741
-
Role of lipopolysaccharides and lipoteichoic acids on C-Chrysophsin-1 interactions with model Gram-positive and Gram-negative bacterial membranes.Biointerphases. 2020 May 26;15(3):031007. doi: 10.1116/1.5130774. Biointerphases. 2020. PMID: 32456440
-
NMR Structures and Interactions of Antimicrobial Peptides with Lipopolysaccharide: Connecting Structures to Functions.Curr Top Med Chem. 2016;16(1):4-15. doi: 10.2174/1568026615666150703121943. Curr Top Med Chem. 2016. PMID: 26139110 Review.
-
Structure and mechanism of beta-hairpin antimicrobial peptides in lipid bilayers from solid-state NMR spectroscopy.Mol Biosyst. 2009 Apr;5(4):317-22. doi: 10.1039/b820398a. Epub 2009 Jan 27. Mol Biosyst. 2009. PMID: 19396367 Free PMC article. Review.
Cited by
-
Membrane-Active Epithelial Keratin 6A Fragments (KAMPs) Are Unique Human Antimicrobial Peptides with a Non-αβ Structure.Front Microbiol. 2016 Nov 11;7:1799. doi: 10.3389/fmicb.2016.01799. eCollection 2016. Front Microbiol. 2016. PMID: 27891122 Free PMC article.
-
Conformational disorder of membrane peptides investigated from solid-state NMR line widths and line shapes.J Phys Chem B. 2011 Sep 15;115(36):10758-67. doi: 10.1021/jp205002n. Epub 2011 Aug 18. J Phys Chem B. 2011. PMID: 21806038 Free PMC article.
-
Atomic-Resolution Structures and Mode of Action of Clinically Relevant Antimicrobial Peptides.Int J Mol Sci. 2022 Apr 20;23(9):4558. doi: 10.3390/ijms23094558. Int J Mol Sci. 2022. PMID: 35562950 Free PMC article. Review.
-
Synthetic molecular evolution of pore-forming peptides by iterative combinatorial library screening.ACS Chem Biol. 2013 Apr 19;8(4):823-31. doi: 10.1021/cb300598k. Epub 2013 Feb 20. ACS Chem Biol. 2013. PMID: 23394375 Free PMC article.
-
Protegrin-1 and Analogues Against Acinetobacter baumannii: A Narrative Review.Pharmaceuticals (Basel). 2025 Feb 20;18(3):289. doi: 10.3390/ph18030289. Pharmaceuticals (Basel). 2025. PMID: 40143068 Free PMC article. Review.
References
-
- Levy SB, Marshall B. Antibacterial resistance worldwide: causes, challenges and responses. Nat Med. 2004;10:S122–S129. - PubMed
-
- Hancock RE, Sahl HG. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat Biotechnol. 2006;24:1551–1557. - PubMed
-
- Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. - PubMed
-
- Brogden KA. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol. 2005;3:238–250. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

