Integration of lipidomics and transcriptomics data towards a systems biology model of sphingolipid metabolism
- PMID: 21303545
- PMCID: PMC3047436
- DOI: 10.1186/1752-0509-5-26
Integration of lipidomics and transcriptomics data towards a systems biology model of sphingolipid metabolism
Abstract
Background: Sphingolipids play important roles in cell structure and function as well as in the pathophysiology of many diseases. Many of the intermediates of sphingolipid biosynthesis are highly bioactive and sometimes have antagonistic activities, for example, ceramide promotes apoptosis whereas sphingosine-1-phosphate can inhibit apoptosis and induce cell growth; therefore, quantification of the metabolites and modeling of the sphingolipid network is imperative for an understanding of sphingolipid biology.
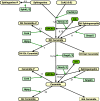
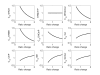
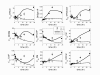
Results: In this direction, the LIPID MAPS Consortium is developing methods to quantitate the sphingolipid metabolites in mammalian cells and is investigating their application to studies of the activation of the RAW264.7 macrophage cell by a chemically defined endotoxin, Kdo2-Lipid A. Herein, we describe a model for the C16-branch of sphingolipid metabolism (i.e., for ceramides with palmitate as the N-acyl-linked fatty acid, which is selected because it is a major subspecies for all categories of complex sphingolipids in RAW264.7 cells) integrating lipidomics and transcriptomics data and using a two-step matrix-based approach to estimate the rate constants from experimental data. The rate constants obtained from the first step are further refined using generalized constrained nonlinear optimization. The resulting model fits the experimental data for all species. The robustness of the model is validated through parametric sensitivity analysis.
Conclusions: A quantitative model of the sphigolipid pathway is developed by integrating metabolomics and transcriptomics data with legacy knowledge. The model could be used to design experimental studies of how genetic and pharmacological perturbations alter the flux through this important lipid biosynthetic pathway.
Figures


References
-
- Zheng W, Kollmeyer J, Symolon H, Momin A, Munter E, Wang E, Kelly S, Allegood JC, Liu Y, Peng Q. et al. Ceramides and other bioactive sphingolipid backbones in health and disease: lipidomic analysis, metabolism and roles in membrane structure, dynamics, signaling and autophagy. Biochim Biophys Acta. 2006;1758(12):1864–1884. doi: 10.1016/j.bbamem.2006.08.009. - DOI - PubMed

