Translational recoding as a feedback controller: systems approaches reveal polyamine-specific effects on the antizyme ribosomal frameshift
- PMID: 21303766
- PMCID: PMC3113565
- DOI: 10.1093/nar/gkq1349
Translational recoding as a feedback controller: systems approaches reveal polyamine-specific effects on the antizyme ribosomal frameshift
Abstract
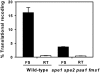
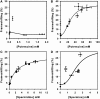
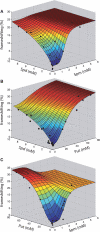
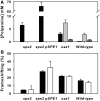
The antizyme protein, Oaz1, regulates synthesis of the polyamines putrescine, spermidine and spermine by controlling stability of the polyamine biosynthetic enzyme, ornithine decarboxylase. Antizyme mRNA translation depends upon a polyamine-stimulated +1 ribosomal frameshift, forming a complex negative feedback system in which the translational frameshifting event may be viewed in engineering terms as a feedback controller for intracellular polyamine concentrations. In this article, we present the first systems level study of the characteristics of this feedback controller, using an integrated experimental and modeling approach. Quantitative analysis of mutant yeast strains in which polyamine synthesis and interconversion were blocked revealed marked variations in frameshift responses to the different polyamines. Putrescine and spermine, but not spermidine, showed evidence of co-operative stimulation of frameshifting and the existence of multiple ribosome binding sites. Combinatorial polyamine treatments showed polyamines compete for binding to common ribosome sites. Using concepts from enzyme kinetics and control engineering, a mathematical model of the translational controller was developed to describe these complex ribosomal responses to combinatorial polyamine effects. Each one of a range of model predictions was successfully validated against experimental frameshift frequencies measured in S-adenosylmethionine-decarboxylase and antizyme mutants, as well as in the wild-type genetic background.
Figures

Similar articles
-
Polyamine sensing by nascent ornithine decarboxylase antizyme stimulates decoding of its mRNA.Nature. 2011 Sep 7;477(7365):490-4. doi: 10.1038/nature10393. Nature. 2011. PMID: 21900894
-
Hypusinated eIF5A Promotes Ribosomal Frameshifting during Decoding of ODC Antizyme mRNA in Saccharomyces cerevisiae.Int J Mol Sci. 2022 Oct 26;23(21):12972. doi: 10.3390/ijms232112972. Int J Mol Sci. 2022. PMID: 36361762 Free PMC article.
-
Conservation of polyamine regulation by translational frameshifting from yeast to mammals.EMBO J. 2000 Apr 17;19(8):1907-17. doi: 10.1093/emboj/19.8.1907. EMBO J. 2000. PMID: 10775274 Free PMC article.
-
The antizyme family for regulating polyamines.J Biol Chem. 2018 Nov 30;293(48):18730-18735. doi: 10.1074/jbc.TM118.003339. Epub 2018 Oct 24. J Biol Chem. 2018. PMID: 30355739 Free PMC article. Review.
-
Polyamine homoeostasis.Essays Biochem. 2009 Nov 4;46:11-24. doi: 10.1042/bse0460002. Essays Biochem. 2009. PMID: 20095967 Review.
Cited by
-
Obesity-Associated Colorectal Cancer.Int J Mol Sci. 2024 Aug 14;25(16):8836. doi: 10.3390/ijms25168836. Int J Mol Sci. 2024. PMID: 39201522 Free PMC article. Review.
-
Pan-cancer analysis of transcriptional metabolic dysregulation using The Cancer Genome Atlas.Nat Commun. 2018 Dec 14;9(1):5330. doi: 10.1038/s41467-018-07232-8. Nat Commun. 2018. PMID: 30552315 Free PMC article.
-
Spermidine, a sensor for antizyme 1 expression regulates intracellular polyamine homeostasis.Amino Acids. 2014 Aug;46(8):2005-13. doi: 10.1007/s00726-014-1757-4. Epub 2014 May 14. Amino Acids. 2014. PMID: 24824458 Free PMC article.
-
Depletion of SAM leading to loss of heterochromatin drives muscle stem cell ageing.Nat Metab. 2024 Jan;6(1):153-168. doi: 10.1038/s42255-023-00955-z. Epub 2024 Jan 19. Nat Metab. 2024. PMID: 38243132 Free PMC article.
-
Role of Polyamines and Hypusine in β Cells and Diabetes Pathogenesis.Metabolites. 2022 Apr 12;12(4):344. doi: 10.3390/metabo12040344. Metabolites. 2022. PMID: 35448531 Free PMC article. Review.
References
-
- Wallace HM, Fraser AV. Inhibitors of polyamine metabolism. Amino Acids. 2004;26:353–365. - PubMed
-
- Poulin R, Coward JK, Lakanen JR, Pegg AE. Enhancement of the spermidine uptake system and lethal effects of spermidine overaccumulation in ornithine decarboxylase-overproducing L1210 cells under hyposmotic stress. J. Biol. Chem. 1993;268:4690–4698. - PubMed
-
- Tobias KE, Kahana C. Exposure to ornithine results in excessive accumulation of putrescine and apoptotic cell death in ornithine decarboxylase overproducing mouse myeloma cells. Cell Growth Differ. 1995;6:1279–1285. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

