ORS1, an H₂O₂-responsive NAC transcription factor, controls senescence in Arabidopsis thaliana
- PMID: 21303842
- PMCID: PMC3063519
- DOI: 10.1093/mp/ssq080
ORS1, an H₂O₂-responsive NAC transcription factor, controls senescence in Arabidopsis thaliana
Abstract
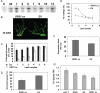
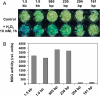
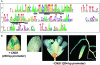
We report here that ORS1, a previously uncharacterized member of the NAC transcription factor family, controls leaf senescence in Arabidopsis thaliana. Overexpression of ORS1 accelerates senescence in transgenic plants, whereas its inhibition delays it. Genes acting downstream of ORS1 were identified by global expression analysis using transgenic plants producing dexamethasone-inducible ORS1-GR fusion protein. Of the 42 up-regulated genes, 30 (~70%) were previously shown to be up-regulated during age-dependent senescence. We also observed that 32 (~76%) of the ORS1-dependent genes were induced by long-term (4 d), but not short-term (6 h) salinity stress (150 mM NaCl). Furthermore, expression of 16 and 24 genes, respectively, was induced after 1 and 5 h of treatment with hydrogen peroxide (H₂O₂), a reactive oxygen species known to accumulate during salinity stress. ORS1 itself was found to be rapidly and strongly induced by H₂O₂ treatment in both leaves and roots. Using in vitro binding site selection, we determined the preferred binding motif of ORS1 and found it to be present in half of the ORS1-dependent genes. ORS1 is a paralog of ORE1/ANAC092/AtNAC2, a previously reported regulator of leaf senescence. Phylogenetic footprinting revealed evolutionary conservation of the ORS1 and ORE1 promoter sequences in different Brassicaceae species, indicating strong positive selection acting on both genes. We conclude that ORS1, similarly to ORE1, triggers expression of senescence-associated genes through a regulatory network that may involve cross-talk with salt- and H₂O₂-dependent signaling pathways.
Figures



References
-
- Agüera E, Cabello P, de la Haba P. Induction of leaf senescence by low nitrogen nutrition in sunflower (Helianthus annuus) plants. Physiol Plant. 2010;138:256–267. - PubMed
-
- Albacete A, Martínez-Andújar C, Ghanem ME, Acosta M, Sánchez-Bravo J, Asins MJ, Cuartero J, Lutts S, Dodd IC, Pérez-Alfocea F. Rootstock-mediated changes in xylem ionic and hormonal status are correlated with delayed leaf senescence, and increased leaf area and crop productivity in salinized tomato. Plant Cell Environ. 2009;32:928–938. - PubMed
-
- Ay N, Irmler K, Fischer A, Uhlemann R, Reuter G, Humbeck K. Epigenetic programming via histone methylation at WRKY53 controls leaf senescence in Arabidopsis thaliana. Plant J. 2009;58:333–346. - PubMed
-
- Balazadeh S, Parlitz S, Mueller-Roeber B, Meyer RC. Natural variation for developmental leaf and plant senescence in Arabidopsis thaliana. Plant Biol. 2008a;(Suppl 1):136–147. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

