Characterization of a Dchs1 mutant mouse reveals requirements for Dchs1-Fat4 signaling during mammalian development
- PMID: 21303848
- PMCID: PMC3035097
- DOI: 10.1242/dev.057166
Characterization of a Dchs1 mutant mouse reveals requirements for Dchs1-Fat4 signaling during mammalian development
Abstract
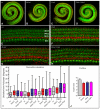
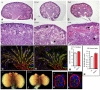
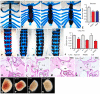
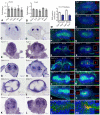
The Drosophila Dachsous and Fat proteins function as ligand and receptor, respectively, for an intercellular signaling pathway that regulates Hippo signaling and planar cell polarity. Although gene-targeted mutations in two mammalian Fat genes have been described, whether mammals have a Fat signaling pathway equivalent to that in Drosophila, and what its biological functions might be, have remained unclear. Here, we describe a gene-targeted mutation in a murine Dachsous homolog, Dchs1. Analysis of the phenotypes of Dchs1 mutant mice and comparisons with Fat4 mutant mice identify requirements for these genes in multiple organs, including the ear, kidney, skeleton, intestine, heart and lung. Dchs1 and Fat4 single mutants and Dchs1 Fat4 double mutants have similar phenotypes throughout the body. In some cases, these phenotypes suggest that Dchs1-Fat4 signaling influences planar cell polarity. In addition to the appearance of cysts in newborn kidneys, we also identify and characterize a requirement for Dchs1 and Fat4 in growth, branching and cell survival during early kidney development. Dchs1 and Fat4 are predominantly expressed in mesenchymal cells in multiple organs, and mutation of either gene increases protein staining for the other. Our analysis implies that Dchs1 and Fat4 function as a ligand-receptor pair during murine development, and identifies novel requirements for Dchs1-Fat4 signaling in multiple organs.
Figures



References
-
- Basson M. A., Watson-Johnson J., Shakya R., Akbulut S., Hyink D., Costantini F. D., Wilson P. D., Mason I. J., Licht J. D. (2006). Branching morphogenesis of the ureteric epithelium during kidney development is coordinated by the opposing functions of GDNF and Sprouty1. Dev. Biol. 299, 466-477 - PubMed
-
- Cardoso W. V., Lü J. (2006). Regulation of early lung morphogenesis: questions, facts and controversies. Development 133, 1611-1624 - PubMed
-
- Casal J., Struhl G., Lawrence P. (2002). Developmental compartments and planar polarity in Drosophila. Curr. Biol. 12, 1189-1198 - PubMed
-
- Castillejo-Lopez C., Arias W. M., Baumgartner S. (2004). The fat-like gene of Drosophila is the true orthologue of vertebrate fat cadherins and is involved in the formation of tubular organs. J. Biol. Chem. 279, 24034-24043 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

