Subunit organization of the human INO80 chromatin remodeling complex: an evolutionarily conserved core complex catalyzes ATP-dependent nucleosome remodeling
- PMID: 21303910
- PMCID: PMC3064184
- DOI: 10.1074/jbc.M111.222505
Subunit organization of the human INO80 chromatin remodeling complex: an evolutionarily conserved core complex catalyzes ATP-dependent nucleosome remodeling
Abstract
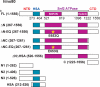
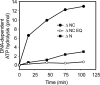
We previously identified and purified a human ATP-dependent chromatin remodeling complex with similarity to the Saccharomyces cerevisiae INO80 complex (Jin, J., Cai, Y., Yao, T., Gottschalk, A. J., Florens, L., Swanson, S. K., Gutierrez, J. L., Coleman, M. K., Workman, J. L., Mushegian, A., Washburn, M. P., Conaway, R. C., and Conaway, J. W. (2005) J. Biol. Chem. 280, 41207-41212) and demonstrated that it is composed of (i) a Snf2 family ATPase (hIno80) related in sequence to the S. cerevisiae Ino80 ATPase; (ii) seven additional evolutionarily conserved subunits orthologous to yeast INO80 complex subunits; and (iii) six apparently metazoan-specific subunits. In this report, we present evidence that the human INO80 complex is composed of three modules that assemble with three distinct domains of the hIno80 ATPase. These modules include (i) one that is composed of the N terminus of the hIno80 protein and all of the metazoan-specific subunits and is not required for ATP-dependent nucleosome remodeling; (ii) a second that is composed of the hIno80 Snf2-like ATPase/helicase and helicase-SANT-associated/post-HSA (HSA/PTH) domain, the actin-related proteins Arp4 and Arp8, and the GLI-Kruppel family transcription factor YY1; and (iii) a third that is composed of the hIno80 Snf2 ATPase domain, the Ies2 and Ies6 proteins, the AAA(+) ATPases Tip49a and Tip49b, and the actin-related protein Arp5. Through purification and characterization of hINO80 complex subassemblies, we demonstrate that ATP-dependent nucleosome remodeling by the hINO80 complex is catalyzed by a core complex comprising the hIno80 protein HSA/PTH and Snf2 ATPase domains acting in concert with YY1 and the complete set of its evolutionarily conserved subunits. Taken together, our findings shed new light on the structure and function of the INO80 chromatin-remodeling complex.
Figures


References
-
- Conaway R. C., Conaway J. W. (2009) Trends Biochem. Sci. 34, 71–77 - PubMed
-
- Ebbert R., Birkmann A., Schüller H. J. (1999) Mol. Microbiol. 32, 741–751 - PubMed
-
- Shen X., Mizuguchi G., Hamiche A., Wu C. (2000) Nature 406, 541–544 - PubMed
-
- Jin J., Cai Y., Yao T., Gottschalk A. J., Florens L., Swanson S. K., Gutiérrez J. L., Coleman M. K., Workman J. L., Mushegian A., Washburn M. P., Conaway R. C., Conaway J. W. (2005) J. Biol. Chem. 280, 41207–41212 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

