Replication and functional genomic analyses of the breast cancer susceptibility locus at 6q25.1 generalize its importance in women of chinese, Japanese, and European ancestry
- PMID: 21303983
- PMCID: PMC3083305
- DOI: 10.1158/0008-5472.CAN-10-2733
Replication and functional genomic analyses of the breast cancer susceptibility locus at 6q25.1 generalize its importance in women of chinese, Japanese, and European ancestry
Abstract
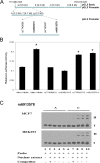
We evaluated the generalizability of a single nucleotide polymorphism (SNP), rs2046210 (A/G allele), associated with breast cancer risk that was initially identified at 6q25.1 in a genome-wide association study conducted among Chinese women. In a pooled analysis of more than 31,000 women of East-Asian, European, and African ancestry, we found a positive association for rs2046210 and breast cancer risk in Chinese women [ORs (95% CI) = 1.30 (1.22-1.38) and 1.64 (1.50-1.80) for the AG and AA genotypes, respectively, P for trend = 1.54 × 10⁻³⁰], Japanese women [ORs (95% CI) = 1.31 (1.13-1.52) and 1.37 (1.06-1.76), P for trend = 2.51 × 10⁻⁴], and European-ancestry American women [ORs (95% CI) = 1.07 (0.99-1.16) and 1.18 (1.04-1.34), P for trend = 0.0069]. No association with this SNP, however, was observed in African American women [ORs (95% CI) = 0.81 (0.63-1.06) and 0.85 (0.65-1.11) for the AG and AA genotypes, respectively, P for trend = 0.4027]. In vitro functional genomic studies identified a putative functional variant, rs6913578. This SNP is 1,440 bp downstream of rs2046210 and is in high linkage disequilibrium with rs2046210 in Chinese (r(2) = 0.91) and European-ancestry (r² = 0.83) populations, but not in Africans (r² = 0.57). SNP rs6913578 was found to be associated with breast cancer risk in Chinese and European-ancestry American women. After adjusting for rs2046210, the association of rs6913578 with breast cancer risk in African Americans approached borderline significance. Results from this large consortium study confirmed the association of rs2046210 with breast cancer risk among women of Chinese, Japanese, and European ancestry. This association may be explained in part by a putatively functional variant (rs6913578) identified in the region.
©2011 AACR.
Figures

References
-
- Nathanson KL, Wooster R, Weber BL. Breast cancer genetics: what we know and what we need. Nat Med. 2001 May;7(5):552–6. - PubMed
-
- Balmain A, Gray J, Ponder B. The genetics and genomics of cancer. Nat Genet. 2003 Mar;33(Suppl):238–44. - PubMed
-
- Gao YT, Shu XO, Dai Q, Potter JD, Brinton LA, Wen W, et al. Association of menstrual and reproductive factors with breast cancer risk: results from the Shanghai Breast Cancer Study. Int J Cancer. 2000;87(2):295–300. - PubMed
-
- Zhang L, Gu L, Qian B, Hao X, Zhang W, Wei Q, et al. Association of genetic polymorphisms of ER-alpha and the estradiol-synthesizing enzyme genes CYP17 and CYP19 with breast cancer risk in Chinese women. Breast Cancer Res Treat. 2009 Mar;114(2):327–38. - PubMed
Publication types
MeSH terms
Grants and funding
- CA54281/CA/NCI NIH HHS/United States
- P30 ES010126/ES/NIEHS NIH HHS/United States
- P30CA68485/CA/NCI NIH HHS/United States
- R01CA64277/CA/NCI NIH HHS/United States
- R01CA92585/CA/NCI NIH HHS/United States
- R01 CA092447/CA/NCI NIH HHS/United States
- R01 CA092585/CA/NCI NIH HHS/United States
- CA63464/CA/NCI NIH HHS/United States
- R01 CA047147/CA/NCI NIH HHS/United States
- R01 CA069664/CA/NCI NIH HHS/United States
- R01CA124558/CA/NCI NIH HHS/United States
- P30ES010126/ES/NIEHS NIH HHS/United States
- R01 CA047305/CA/NCI NIH HHS/United States
- P30 ES009089/ES/NIEHS NIH HHS/United States
- R01 CA105197/CA/NCI NIH HHS/United States
- R01 CA100374/CA/NCI NIH HHS/United States
- R01CA100374/CA/NCI NIH HHS/United States
- R01 CA064277/CA/NCI NIH HHS/United States
- P30ES009089/ES/NIEHS NIH HHS/United States
- R01 CA118229/CA/NCI NIH HHS/United States
- R01 CA124558/CA/NCI NIH HHS/United States
- R01CA118229/CA/NCI NIH HHS/United States
- R01 CA063464/CA/NCI NIH HHS/United States
- R01 CA054281/CA/NCI NIH HHS/United States
- U01 CA063464/CA/NCI NIH HHS/United States
- R01 CA132839/CA/NCI NIH HHS/United States
- P30 CA068485/CA/NCI NIH HHS/United States
- U01CA/ES66572/CA/NCI NIH HHS/United States
- R01CA092447/CA/NCI NIH HHS/United States
- CA132839/CA/NCI NIH HHS/United States
- R37 CA054281/CA/NCI NIH HHS/United States
- R01 CA47147/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

