The conserved factor DE-ETIOLATED 1 cooperates with CUL4-DDB1DDB2 to maintain genome integrity upon UV stress
- PMID: 21304489
- PMCID: PMC3061027
- DOI: 10.1038/emboj.2011.20
The conserved factor DE-ETIOLATED 1 cooperates with CUL4-DDB1DDB2 to maintain genome integrity upon UV stress
Abstract
Plants and many other eukaryotes can make use of two major pathways to cope with mutagenic effects of light, photoreactivation and nucleotide excision repair (NER). While photoreactivation allows direct repair by photolyase enzymes using light energy, NER requires a stepwise mechanism with several protein complexes acting at the levels of lesion detection, DNA incision and resynthesis. Here we investigated the involvement in NER of DE-ETIOLATED 1 (DET1), an evolutionarily conserved factor that associates with components of the ubiquitylation machinery in plants and mammals and acts as a negative repressor of light-driven photomorphogenic development in Arabidopsis. Evidence is provided that plant DET1 acts with CULLIN4-based ubiquitin E3 ligase, and that appropriate dosage of DET1 protein is necessary for efficient removal of UV photoproducts through the NER pathway. Moreover, DET1 is required for CULLIN4-dependent targeted degradation of the UV-lesion recognition factor DDB2. Finally, DET1 protein is degraded concomitantly with DDB2 upon UV irradiation in a CUL4-dependent mechanism. Altogether, these data suggest that DET1 and DDB2 cooperate during the excision repair process.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
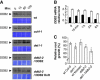
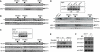

Similar articles
-
Arabidopsis DDB1-CUL4 E3 ligase complexes in det1 salt/osmotic stress resistant germination.Plant Signal Behav. 2016 Sep;11(9):e1223004. doi: 10.1080/15592324.2016.1223004. Plant Signal Behav. 2016. PMID: 27547879 Free PMC article.
-
Regulation and role of Arabidopsis CUL4-DDB1A-DDB2 in maintaining genome integrity upon UV stress.PLoS Genet. 2008 Jun 13;4(6):e1000093. doi: 10.1371/journal.pgen.1000093. PLoS Genet. 2008. PMID: 18551167 Free PMC article.
-
Arabidopsis DDB1-CUL4 ASSOCIATED FACTOR1 forms a nuclear E3 ubiquitin ligase with DDB1 and CUL4 that is involved in multiple plant developmental processes.Plant Cell. 2008 Jun;20(6):1437-55. doi: 10.1105/tpc.108.058891. Epub 2008 Jun 13. Plant Cell. 2008. PMID: 18552200 Free PMC article.
-
The photomorphogenic repressors COP1 and DET1: 20 years later.Trends Plant Sci. 2012 Oct;17(10):584-93. doi: 10.1016/j.tplants.2012.05.004. Epub 2012 Jun 15. Trends Plant Sci. 2012. PMID: 22705257 Review.
-
Detecting UV-lesions in the genome: The modular CRL4 ubiquitin ligase does it best!FEBS Lett. 2011 Sep 16;585(18):2818-25. doi: 10.1016/j.febslet.2011.04.064. Epub 2011 May 6. FEBS Lett. 2011. PMID: 21550341 Review.
Cited by
-
Crossover localisation is regulated by the neddylation posttranslational regulatory pathway.PLoS Biol. 2014 Aug 12;12(8):e1001930. doi: 10.1371/journal.pbio.1001930. eCollection 2014 Aug. PLoS Biol. 2014. PMID: 25116939 Free PMC article.
-
Combined Multi-Omics and Co-Expression Network Analyses Uncover the Pigment Accumulation Mechanism of Orange-Red Petals in Brassica napus L.Biology (Basel). 2025 Jun 13;14(6):693. doi: 10.3390/biology14060693. Biology (Basel). 2025. PMID: 40563944 Free PMC article.
-
Arabidopsis WD repeat domain55 Interacts with DNA damaged binding protein1 and is required for apical patterning in the embryo.Plant Cell. 2012 Mar;24(3):1013-33. doi: 10.1105/tpc.111.089425. Epub 2012 Mar 23. Plant Cell. 2012. PMID: 22447688 Free PMC article.
-
Arabidopsis CRL4 Complexes: Surveying Chromatin States and Gene Expression.Front Plant Sci. 2019 Sep 17;10:1095. doi: 10.3389/fpls.2019.01095. eCollection 2019. Front Plant Sci. 2019. PMID: 31608079 Free PMC article. Review.
-
A novel cis-acting element from the 3'UTR of DNA damage-binding protein 2 mRNA links transcriptional and post-transcriptional regulation of gene expression.Nucleic Acids Res. 2013 Jun;41(11):5692-703. doi: 10.1093/nar/gkt279. Epub 2013 Apr 19. Nucleic Acids Res. 2013. PMID: 23605047 Free PMC article.
References
-
- Al Khateeb WM, Schroeder DF (2009) Overexpression of Arabidopsis damaged DNA binding protein 1A (DDB1A) enhances UV tolerance. Plant Mol Biol 70: 371–383 - PubMed
-
- Angers S, Li T, Yi X, MacCoss MJ, Moon RT, Zheng N (2006) Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature 443: 590–593 - PubMed
-
- Benvenuto G, Formiggini F, Laflamme P, Malakhov M, Bowler C (2002) The photomorphogenesis regulator DET1 binds the amino-terminal tail of histone H2B in a nucleosome context. Curr Biol 12: 1529–1534 - PubMed
-
- Bernhardt A, Lechner E, Hano P, Schade V, Dieterle M, Anders M, Dubin MJ, Benvenuto G, Bowler C, Genschik P, Hellmann H (2006) CUL4 associates with DDB1 and DET1 and its downregulation affects diverse aspects of development in Arabidopsis thaliana. Plant J 47: 591–603 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

