Transcription reprogramming during root nodule development in Medicago truncatula
- PMID: 21304580
- PMCID: PMC3029352
- DOI: 10.1371/journal.pone.0016463
Transcription reprogramming during root nodule development in Medicago truncatula
Abstract
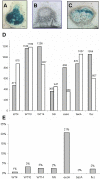
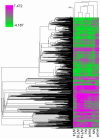
Many genes which are associated with root nodule development and activity in the model legume Medicago truncatula have been described. However information on precise stages of activation of these genes and their corresponding transcriptional regulators is often lacking. Whether these regulators are shared with other plant developmental programs also remains an open question. Here detailed microarray analyses have been used to study the transcriptome of root nodules induced by either wild type or mutant strains of Sinorhizobium meliloti. In this way we have defined eight major activation patterns in nodules and identified associated potential regulatory genes. We have shown that transcription reprogramming during consecutive stages of nodule differentiation occurs in four major phases, respectively associated with (i) early signalling events and/or bacterial infection; plant cell differentiation that is either (ii) independent or (iii) dependent on bacteroid differentiation; (iv) nitrogen fixation. Differential expression of several genes involved in cytokinin biosynthesis was observed in early symbiotic nodule zones, suggesting that cytokinin levels are actively controlled in this region. Taking advantage of databases recently developed for M. truncatula, we identified a small subset of gene expression regulators that were exclusively or predominantly expressed in nodules, whereas most other regulators were also activated under other conditions, and notably in response to abiotic or biotic stresses. We found evidence suggesting the activation of the jasmonate pathway in both wild type and mutant nodules, thus raising questions about the role of jasmonate during nodule development. Finally, quantitative RT-PCR was used to analyse the expression of a series of nodule regulator and marker genes at early symbiotic stages in roots and allowed us to distinguish several early stages of gene expression activation or repression.
Conflict of interest statement
Figures





References
-
- Riechmann JL, Heard J, Martin G, Reuber L, Jiang C, et al. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science. 2000;290:2105–2110. - PubMed
-
- Qu LJ, Zhu YX. Transcription factor families in Arabidopsis: major progress and outstanding issues for future research. Curr Opin Plant Biol. 2006;9:544–549. - PubMed
-
- Oldroyd GE, Downie JA. Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu Rev Plant Biol. 2008;59:519–546. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

