Complete genome sequence of Capnocytophaga ochracea type strain (VPI 2845)
- PMID: 21304645
- PMCID: PMC3035226
- DOI: 10.4056/sigs.15195
Complete genome sequence of Capnocytophaga ochracea type strain (VPI 2845)
Abstract
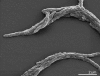
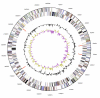
Capnocytophaga ochracea (Prévot et al. 1956) Leadbetter et al. 1982 is the type species of the genus Capnocytophaga. It is of interest because of its location in the Flavobacteriaceae, a genomically not yet charted family within the order Flavobacteriales. The species grows as fusiform to rod shaped cells which tend to form clumps and are able to move by gliding. C. ochracea is known as a capnophilic (CO(2)-requiring) organism with the ability to grow under anaerobic as well as aerobic conditions (oxygen concentration larger than 15%), here only in the presence of 5% CO(2). Strain VPI 2845(T), the type strain of the species, is portrayed in this report as a gliding, Gram-negative bacterium, originally isolated from a human oral cavity. Here we describe the features of this organism, together with the complete genome sequence, and annotation. This is the first completed genome sequence from the flavobacterial genus Capnocytophaga, and the 2,612,925 bp long single replicon genome with its 2193 protein-coding and 59 RNA genes is a part of the Genomic Encyclopedia of Bacteria and Archaea project.
Keywords: Flavobacteriaceae; capnophilic; gingivitis; gliding; periodontitis.
Figures

Similar articles
-
Complete genome sequence of Brachybacterium faecium type strain (Schefferle 6-10).Stand Genomic Sci. 2009 Jul 20;1(1):3-11. doi: 10.4056/sigs.492. Stand Genomic Sci. 2009. PMID: 21304631 Free PMC article.
-
Complete genome sequence of Slackia heliotrinireducens type strain (RHS 1).Stand Genomic Sci. 2009 Nov 22;1(3):234-41. doi: 10.4056/sigs.37633. Stand Genomic Sci. 2009. PMID: 21304663 Free PMC article.
-
Complete genome sequence of Eggerthella lenta type strain (IPP VPI 0255).Stand Genomic Sci. 2009 Sep 28;1(2):174-82. doi: 10.4056/sigs.33592. Stand Genomic Sci. 2009. PMID: 21304654 Free PMC article.
-
Complete genome sequence of Catenulispora acidiphila type strain (ID 139908).Stand Genomic Sci. 2009 Sep 24;1(2):119-25. doi: 10.4056/sigs.17259. Stand Genomic Sci. 2009. PMID: 21304647 Free PMC article.
-
Complete genome sequence of Atopobium parvulum type strain (IPP 1246).Stand Genomic Sci. 2009 Sep 23;1(2):166-73. doi: 10.4056/sigs.29547. Stand Genomic Sci. 2009. PMID: 21304653 Free PMC article.
Cited by
-
Involvement of the Type IX Secretion System in Capnocytophaga ochracea Gliding Motility and Biofilm Formation.Appl Environ Microbiol. 2016 Jan 4;82(6):1756-1766. doi: 10.1128/AEM.03452-15. Appl Environ Microbiol. 2016. PMID: 26729712 Free PMC article.
-
Genome sequence of the Antarctic rhodopsins-containing flavobacterium Gillisia limnaea type strain (R-8282(T)).Stand Genomic Sci. 2012 Oct 10;7(1):107-19. doi: 10.4056/sigs.3216895. Stand Genomic Sci. 2012. PMID: 23450183 Free PMC article.
-
High-quality-draft genome sequence of the yellow-pigmented flavobacterium Joostella marina type strain (En5(T)).Stand Genomic Sci. 2013 Apr 15;8(1):37-46. doi: 10.4056/sigs.3537045. eCollection 2013 Apr 15. Stand Genomic Sci. 2013. PMID: 23961310 Free PMC article.
-
Environmental and gut bacteroidetes: the food connection.Front Microbiol. 2011 May 30;2:93. doi: 10.3389/fmicb.2011.00093. eCollection 2011. Front Microbiol. 2011. PMID: 21747801 Free PMC article.
-
Gliding motility and Por secretion system genes are widespread among members of the phylum bacteroidetes.J Bacteriol. 2013 Jan;195(2):270-8. doi: 10.1128/JB.01962-12. Epub 2012 Nov 2. J Bacteriol. 2013. PMID: 23123910 Free PMC article.
References
-
- de Cadore F, Prévot AR, Tardieux P, Joubert L. Recherches sur Fusiformis nucleatus (Knorr) et son pouvoir pathogène pour l'homme et les animaux. Ann Inst Pasteur (Paris) 1956; 91:787-798 - PubMed
-
- Leadbetter ER. Validation of the Publication of New Names and New Combinations Previously Effectively Published Outside the IJSB: List No. 8. Int J Syst Bacteriol 1982; 32:266-268
-
- Holdeman LV, Moore WEC. Bacteroides Anaerobe Laboratory Manual, Virginia Polytechnic Institute Anaerobe. Blacksburg, VA 1972.
-
- Sebald M. Etudes sur les bactéries anaérobies gram-négatives asporulée. Lavel, France: Imprimerie Barnéoud S. A.; 1962.
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

