Complete genome sequence of Aminobacterium colombiense type strain (ALA-1)
- PMID: 21304712
- PMCID: PMC3035294
- DOI: 10.4056/sigs.902116
Complete genome sequence of Aminobacterium colombiense type strain (ALA-1)
Abstract
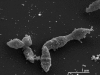
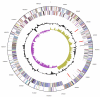
Aminobacterium colombiense Baena et al. 1999 is the type species of the genus Aminobacterium. This genus is of large interest because of its isolated phylogenetic location in the family Synergistaceae, its strictly anaerobic lifestyle, and its ability to grow by fermentation of a limited range of amino acids but not carbohydrates. Here we describe the features of this organism, together with the complete genome sequence and annotation. This is the second completed genome sequence of a member of the family Synergistaceae and the first genome sequence of a member of the genus Aminobacterium. The 1,980,592 bp long genome with its 1,914 protein-coding and 56 RNA genes is part of the Genomic Encyclopedia of Bacteria and Archaea project.
Keywords: GEBA; Synergistaceae; fermentation of amino acids; gram-negative firmicute; strictly anaerobic; syntrophic organism.
Figures

Similar articles
-
Complete genome sequence of Syntrophothermus lipocalidus type strain (TGB-C1).Stand Genomic Sci. 2010 Dec 15;3(3):268-75. doi: 10.4056/sigs.1233249. Stand Genomic Sci. 2010. PMID: 21304731 Free PMC article.
-
Complete genome sequence of Thermanaerovibrio acidaminovorans type strain (Su883).Stand Genomic Sci. 2009 Nov 22;1(3):254-61. doi: 10.4056/sigs.40645. Stand Genomic Sci. 2009. PMID: 21304665 Free PMC article.
-
Complete genome sequence of Calditerrivibrio nitroreducens type strain (Yu37-1).Stand Genomic Sci. 2011 Feb 20;4(1):54-62. doi: 10.4056/sigs.1523807. Stand Genomic Sci. 2011. PMID: 21475587 Free PMC article.
-
Complete genome sequence of Brachyspira murdochii type strain (56-150).Stand Genomic Sci. 2010 Jun 15;2(3):260-9. doi: 10.4056/sigs.831993. Stand Genomic Sci. 2010. PMID: 21304710 Free PMC article.
-
Non-contiguous finished genome sequence of Aminomonas paucivorans type strain (GLU-3).Stand Genomic Sci. 2010 Nov 20;3(3):285-93. doi: 10.4056/sigs.1253298. Stand Genomic Sci. 2010. PMID: 21304733 Free PMC article.
Cited by
-
Microbial insights of enhanced anaerobic conversion of syngas into volatile fatty acids by co-fermentation with carbohydrate-rich synthetic wastewater.Biotechnol Biofuels. 2020 Mar 16;13:53. doi: 10.1186/s13068-020-01694-z. eCollection 2020. Biotechnol Biofuels. 2020. PMID: 32190118 Free PMC article.
-
Complete genome sequence of Syntrophothermus lipocalidus type strain (TGB-C1).Stand Genomic Sci. 2010 Dec 15;3(3):268-75. doi: 10.4056/sigs.1233249. Stand Genomic Sci. 2010. PMID: 21304731 Free PMC article.
-
To the Understanding of Catalysis by D-Amino Acid Transaminases: A Case Study of the Enzyme from Aminobacterium colombiense.Molecules. 2023 Feb 23;28(5):2109. doi: 10.3390/molecules28052109. Molecules. 2023. PMID: 36903355 Free PMC article.
-
Metaproteomics-guided selection of targeted enzymes for bioprospecting of mixed microbial communities.Biotechnol Biofuels. 2017 May 16;10:128. doi: 10.1186/s13068-017-0815-z. eCollection 2017. Biotechnol Biofuels. 2017. PMID: 28523076 Free PMC article.
-
Effect of a Profound Feedstock Change on the Structure and Performance of Biogas Microbiomes.Microorganisms. 2020 Jan 25;8(2):169. doi: 10.3390/microorganisms8020169. Microorganisms. 2020. PMID: 31991721 Free PMC article.
References
-
- Validation of publication of new names and new combinations previously effectively published outside the IJSB. Int J Syst Bacteriol 1999; 49:1325-1326 10.1099/00207713-49-4-1325 - DOI
-
- Baena S, Fardeau ML, Labat M, Ollivier B, Garcia JL, Patel BKC. Aminobacterium mobile sp. nov., a new anaerobic amino-acid-degrading bacterium. Int J Syst Evol Microbiol 2000; 50:259-264 - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

