Detection and removal of biases in the analysis of next-generation sequencing reads
- PMID: 21304912
- PMCID: PMC3031631
- DOI: 10.1371/journal.pone.0016685
Detection and removal of biases in the analysis of next-generation sequencing reads
Abstract
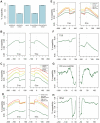
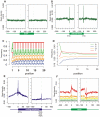
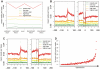
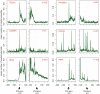
Since the emergence of next-generation sequencing (NGS) technologies, great effort has been put into the development of tools for analysis of the short reads. In parallel, knowledge is increasing regarding biases inherent in these technologies. Here we discuss four different biases we encountered while analyzing various Illumina datasets. These biases are due to both biological and statistical effects that in particular affect comparisons between different genomic regions. Specifically, we encountered biases pertaining to the distributions of nucleotides across sequencing cycles, to mappability, to contamination of pre-mRNA with mRNA, and to non-uniform hydrolysis of RNA. Most of these biases are not specific to one analyzed dataset, but are present across a variety of datasets and within a variety of genomic contexts. Importantly, some of these biases correlated in a highly significant manner with biological features, including transcript length, gene expression levels, conservation levels, and exon-intron architecture, misleadingly increasing the credibility of results due to them. We also demonstrate the relevance of these biases in the context of analyzing an NGS dataset mapping transcriptionally engaged RNA polymerase II (RNAPII) in the context of exon-intron architecture, and show that elimination of these biases is crucial for avoiding erroneous interpretation of the data. Collectively, our results highlight several important pitfalls, challenges and approaches in the analysis of NGS reads.
Conflict of interest statement
Figures


References
-
- Metzker ML. Sequencing technologies - the next generation. Nat Rev Genet. 11:31–46. - PubMed
-
- Flicek P, Birney E. Sense from sequence reads: methods for alignment and assembly. Nat Methods. 2009;6:S6–S12. - PubMed
-
- Medvedev P, Stanciu M, Brudno M. Computational methods for discovering structural variation with next-generation sequencing. Nat Methods. 2009;6:S13–20. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

