Within-host dynamics of the hepatitis C virus quasispecies population in HIV-1/HCV coinfected patients
- PMID: 21304985
- PMCID: PMC3031583
- DOI: 10.1371/journal.pone.0016551
Within-host dynamics of the hepatitis C virus quasispecies population in HIV-1/HCV coinfected patients
Abstract
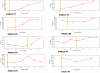
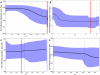
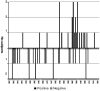
HIV/HCV coinfected individuals under highly active antiretroviral therapy (HAART) represent an interesting model for the investigation of the role played by the immune system in driving the evolution of the HCV quasispecies. We prospectively studied the intra-host evolution of the HCV heterogeneity in 8 coinfected subjects, selected from a cohort of 32 patients initiating HAART: 5 immunological responders (group A) and 3 immunological non-responders (group B), and in two HCV singly infected controls not assuming drugs (group C). For all these subjects at least two serial samples obtained at the first observation (before HAART) and more than 1 year later, underwent clonal sequence analysis of partial E1/E2 sequences, encompassing the whole HVR1. Evolutionary rates, dated phylogenies and population dynamics were co-estimated by using a Bayesian Markov Chain Monte Carlo approach, and site specific selection pressures were estimated by maximum likelihood-based methods. The intra-host evolutionary rates of HCV quasispecies was 10 times higher in subjects treated with HAART than in controls without immunodeficiency (1.9 and 2.3 × 10(-3) sub/site/month in group A and B and 0.29 × 10(-3) sub/site/month in group C individuals). The within-host Bayesian Skyline plot analysis showed an exponential growth of the quasispecies populations in immunological responders, coinciding with a peak in CD4 cell counts. On the contrary, quasispecies population remained constant in group B and in group C controls. A significant positive selection pressure was detected in a half of the patients under HAART and in none of the group C controls. Several sites under significant positive selection were described, mainly included in the HVR1. Our data indicate that different forces, in addition to the selection pressure, drive an exceptionally fast evolution of HCV during HAART immune restoration. We hypothesize that an important role is played by the enlargement of the viral replicative space.
Conflict of interest statement
Figures

References
-
- Eigen M. Selforganization of matter and the evolution of biological macromolecules. Naturwissenschaften. 1971;58:465–523. - PubMed
-
- Farci P, Purcell RH. Clinical significance of hepatitis C virus genotypes and quasispecies. Semin Liver Dis. 2000;20:103–126. - PubMed
-
- Pawlotsky JM, Tsakiris L, Roudot-Thoraval F, Pellet C, Stuyver L, et al. Relationship between hepatitis C virus genotypes and sources of infection in patients with chronic hepatitis C. J Infect Dis. 1995;171:1607–1610. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

