Tartronate semialdehyde reductase defines a novel rate-limiting step in assimilation and bioconversion of glycerol in Ustilago maydis
- PMID: 21305026
- PMCID: PMC3031564
- DOI: 10.1371/journal.pone.0016438
Tartronate semialdehyde reductase defines a novel rate-limiting step in assimilation and bioconversion of glycerol in Ustilago maydis
Abstract
Background: Glycerol is a by-product of biodiesel production. Currently, it has limited applications with low bioconversion efficiency to most metabolites reported. This is partly attributed to the poor knowledge on the glycerol metabolic pathway in bacteria and fungi.
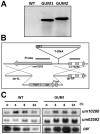
Methodology/principal findings: We have established a fast screening method for identification of genes that improve glycerol utilization in Ustilago maydis. This was done by comparing the growth rates of T-DNA tagged mutant colonies on solid medium using glycerol as the sole carbon source. We present a detailed characterization of one of the mutants, GUM1, which contains a T-DNA element inserted into the promoter region of UM02592 locus (MIPS Ustilago maydis database, MUMDB), leading to enhanced and constitutive expression of its mRNA. We have demonstrated that um02592 encodes a functional tartronate semialdehyde reductase (Tsr1), which showed dual specificity to cofactors NAD(+) and NADP(+) and strong substrate specificity and enantioselectivity for D-glycerate. Improved glycerol assimilation in GUM1 was associated with elevated expression of tsr1 mRNA and this could be phenocopied by over-expression of the gene. Glycolipid accumulation was reduced by 45.2% in the knockout mutant whereas introduction of an extra copy of tsr1 driven by the glyceraldehyde phosphate dehydrogenase promoter increased it by 40.4%.
Conclusions/significance: Our results demonstrate that tartronate semialdehyde reductase (TSR) plays an important role in glycerol assimilation in U. maydis and defines a novel target in genetic engineering for improved conversion of glycerol to higher value products. Our results add significant depth to the understanding of the glycerol metabolic pathway in fungi. We have demonstrated, for the first time, a biological role of a eukaryotic TSR.
Conflict of interest statement
Figures








References
-
- Njau RK, Herndon CA, Hawes JW. New developments in our understanding of the beta-hydroxyacid dehydrogenases. Chem Biol Interact. 2001;130-132:785–791. - PubMed
-
- Hawes JW, Crabb DW, Chan RM, Rougraff PM, Harris RA. Chemical Modification and Site-Directed Mutagenesis Studies of Rat 3-Hydroxyisobutyrate Dehydrogenase. Biochemistry. 1995;34:4231–4237. - PubMed
-
- Njau RK, Herndon CA, Hawes JW. Novel beta-hydroxyacid dehydrogenases in Escherichia coli and Haemophilus influenzae. J Biol Chem. 2000;275:38780–38786. - PubMed
-
- Thompson JC, He BB. Characterization of crude glycerol from biodiesel production from multiple feedstocks. Applied Engineering in Agriculture. 2006;22:261–265.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

