Expression profiling of major histocompatibility and natural killer complex genes reveals candidates for controlling risk of graft versus host disease
- PMID: 21305040
- PMCID: PMC3030590
- DOI: 10.1371/journal.pone.0016582
Expression profiling of major histocompatibility and natural killer complex genes reveals candidates for controlling risk of graft versus host disease
Abstract
Background: The major histocompatibility complex (MHC) is the most important genomic region that contributes to the risk of graft versus host disease (GVHD) after haematopoietic stem cell transplantation. Matching of MHC class I and II genes is essential for the success of transplantation. However, the MHC contains additional genes that also contribute to the risk of developing acute GVHD. It is difficult to identify these genes by genetic association studies alone due to linkage disequilibrium in this region. Therefore, we aimed to identify MHC genes and other genes involved in the pathophysiology of GVHD by mRNA expression profiling.
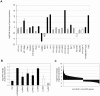
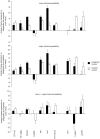
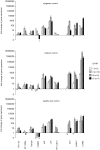
Methodology/principal findings: To reduce the complexity of the task, we used genetically well-defined rat inbred strains and a rat skin explant assay, an in-vitro-model of the graft versus host reaction (GVHR), to analyze the expression of MHC, natural killer complex (NKC), and other genes in cutaneous GVHR. We observed a statistically significant and strong up or down regulation of 11 MHC, 6 NKC, and 168 genes encoded in other genomic regions, i.e. 4.9%, 14.0%, and 2.6% of the tested genes respectively. The regulation of 7 selected MHC and 3 NKC genes was confirmed by quantitative real-time PCR and in independent skin explant assays. In addition, similar regulations of most of the selected genes were observed in GVHD-affected skin lesions of transplanted rats and in human skin explant assays.
Conclusions/significance: We identified rat and human MHC and NKC genes that are regulated during GVHR in skin explant assays and could therefore serve as biomarkers for GVHD. Several of the respective human genes, including HLA-DMB, C2, AIF1, SPR1, UBD, and OLR1, are polymorphic. These candidates may therefore contribute to the genetic risk of GVHD in patients.
Conflict of interest statement
Figures

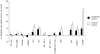


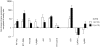
References
-
- Dickinson AM, Middleton PG, Rocha V, Gluckman E, Holler E. Genetic polymorphisms predicting the outcome of bone marrow transplants. Br J Haematol. 2004;127:479–490. - PubMed
-
- Flomenberg N, Baxter-Lowe LA, Confer D, Fernandez-Vina M, Filipovich A, et al. Impact of HLA class I and class II high-resolution matching on outcomes of unrelated donor bone marrow transplantation: HLA-C mismatching is associated with a strong adverse effect on transplantation outcome. Blood. 2004;104:1923–1930. - PubMed
-
- Dickinson AM. Non-HLA genetics and predicting outcome in HSCT. Int J Immunogenet. 2008;35:375–380. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

