MicroRNAs profiling in murine models of acute and chronic asthma: a relationship with mRNAs targets
- PMID: 21305051
- PMCID: PMC3030602
- DOI: 10.1371/journal.pone.0016509
MicroRNAs profiling in murine models of acute and chronic asthma: a relationship with mRNAs targets
Abstract
Background: miRNAs are now recognized as key regulator elements in gene expression. Although they have been associated with a number of human diseases, their implication in acute and chronic asthma and their association with lung remodelling have never been thoroughly investigated.
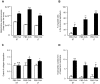
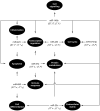
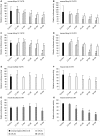
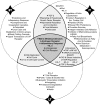
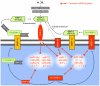
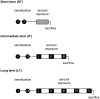
Methodology/principal findings: In order to establish a miRNAs expression profile in lung tissue, mice were sensitized and challenged with ovalbumin mimicking acute, intermediate and chronic human asthma. Levels of lung miRNAs were profiled by microarray and in silico analyses were performed to identify potential mRNA targets and to point out signalling pathways and biological processes regulated by miRNA-dependent mechanisms. Fifty-eight, 66 and 75 miRNAs were found to be significantly modulated at short-, intermediate- and long-term challenge, respectively. Inverse correlation with the expression of potential mRNA targets identified mmu-miR-146b, -223, -29b, -29c, -483, -574-5p, -672 and -690 as the best candidates for an active implication in asthma pathogenesis. A functional validation assay was performed by cotransfecting in human lung fibroblasts (WI26) synthetic miRNAs and engineered expression constructs containing the coding sequence of luciferase upstream of the 3'UTR of various potential mRNA targets. The bioinformatics analysis identified miRNA-linked regulation of several signalling pathways, as matrix metalloproteinases, inflammatory response and TGF-β signalling, and biological processes, including apoptosis and inflammation.
Conclusions/significance: This study highlights that specific miRNAs are likely to be involved in asthma disease and could represent a valuable resource both for biological makers identification and for unveiling mechanisms underlying the pathogenesis of asthma.
Conflict of interest statement
Figures




References
-
- Lange P, Parner J, Vestbo J, Schnohr P, Jensen G. A 15-year follow-up study of ventilatory function in adults with asthma. N Engl J Med. 1998;339:1194–1200. - PubMed
-
- Pascual RM, Peters SP. Airway remodeling contributes to the progressive loss of lung function in asthma: an overview. J Allergy Clin Immunol. 2005;116:477–486; quiz 487. - PubMed
-
- Hirst SJ, Martin JG, Bonacci JV, Chan V, Fixman ED, et al. Proliferative aspects of airway smooth muscle. J Allergy Clin Immunol. 2004;114:S2–17. - PubMed
-
- Di Valentin E, Crahay C, Garbacki N, Hennuy B, Gueders M, et al. New asthma biomarkers: lessons from murine models of acute and chronic asthma. Am J Physiol Lung Cell Mol Physiol. 2009;296:L185–197. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

