Binding between a distal C-terminus fragment of cannabinoid receptor 1 and arrestin-2
- PMID: 21306178
- PMCID: PMC3634367
- DOI: 10.1021/bi1018144
Binding between a distal C-terminus fragment of cannabinoid receptor 1 and arrestin-2
Abstract
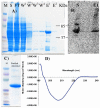
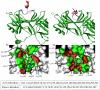
Internalization of G-protein-coupled receptors is mediated by phosphorylation of the C-terminus, followed by binding with the cytosolic protein arrestin. To explore structural factors that may play a role in internalization of cannabinoid receptor 1 (CB1), we utilize a phosphorylated peptide derived from the distal C-terminus of CB1 (CB1(5P)(454-473)). Complexes formed between the peptide and human arrestin-2 (wt-arr2(1-418)) were compared to those formed with a truncated arrestin-2 mutant (tr-arr2(1-382)) using isothermal titration calorimetry and nuclear magnetic resonance spectroscopy. The pentaphosphopeptide CB1(5P)(454-473) adopts a helix-loop conformation, whether binding to full-length arrestin-2 or its truncated mutant. This structure is similar to that of a heptaphosphopeptide, mimicking the distal segment of the rhodopsin C-tail (Rh(7P)(330-348)), binding to visual arrestin, suggesting that this adopted structure bears functional significance. Isothermal titration calorimetry (ITC) experiments show that the CB1(5P)(454-473) peptide binds to tr-arr2(1-382) with higher affinity than to the full-length wt-arr2(1-418). As the observed structure of the bound peptides is similar in either case, we attribute the increased affinity to a more exposed binding site on the N-domain of the truncated arrestin construct. The transferred NOE data from the bound phosphopeptides are used to predict a model describing the interaction with arrestin, using the data driven HADDOCK docking program. The truncation of arrestin-2 provides scope for positively charged residues in the polar core of the protein to interact with phosphates present in the loop of the CB1(5P)(454-473) peptide.
Figures





Similar articles
-
Interaction of a fragment of the cannabinoid CB1 receptor C-terminus with arrestin-2.FEBS Lett. 2007 Oct 16;581(25):5009-16. doi: 10.1016/j.febslet.2007.09.030. Epub 2007 Sep 24. FEBS Lett. 2007. PMID: 17910957 Free PMC article.
-
Cannabinoid Receptor Interacting Protein 1a Competition with β-Arrestin for CB1 Receptor Binding Sites.Mol Pharmacol. 2017 Feb;91(2):75-86. doi: 10.1124/mol.116.104638. Epub 2016 Nov 28. Mol Pharmacol. 2017. PMID: 27895162 Free PMC article.
-
Distinct roles of β-arrestin 1 and β-arrestin 2 in ORG27569-induced biased signaling and internalization of the cannabinoid receptor 1 (CB1).J Biol Chem. 2013 Apr 5;288(14):9790-9800. doi: 10.1074/jbc.M112.438804. Epub 2013 Feb 28. J Biol Chem. 2013. PMID: 23449980 Free PMC article.
-
The cannabinoid type-1 receptor carboxyl-terminus, more than just a tail.J Neurochem. 2011 Apr;117(1):1-18. doi: 10.1111/j.1471-4159.2011.07186.x. Epub 2011 Feb 11. J Neurochem. 2011. PMID: 21244428 Free PMC article. Review.
-
Cannabinoid Receptor Interacting Protein 1a (CRIP1a): Function and Structure.Molecules. 2019 Oct 12;24(20):3672. doi: 10.3390/molecules24203672. Molecules. 2019. PMID: 31614728 Free PMC article. Review.
Cited by
-
Protein Interactors and Trafficking Pathways That Regulate the Cannabinoid Type 1 Receptor (CB1R).Front Mol Neurosci. 2020 Jun 12;13:108. doi: 10.3389/fnmol.2020.00108. eCollection 2020. Front Mol Neurosci. 2020. PMID: 32595453 Free PMC article. Review.
-
Type 1 cannabinoid receptor ligands display functional selectivity in a cell culture model of striatal medium spiny projection neurons.J Biol Chem. 2014 Sep 5;289(36):24845-62. doi: 10.1074/jbc.M114.557025. Epub 2014 Jul 18. J Biol Chem. 2014. PMID: 25037227 Free PMC article.
-
Molecular Interaction between Distal C-Terminal Domain of the CB1 Cannabinoid Receptor and Cannabinoid Receptor Interacting Proteins (CRIP1a/CRIP1b).J Chem Inf Model. 2019 Dec 23;59(12):5294-5303. doi: 10.1021/acs.jcim.9b00948. Epub 2019 Dec 10. J Chem Inf Model. 2019. PMID: 31769975 Free PMC article.
-
Cannabinoid CB1 and CB2 Receptor Signaling and Bias.Cannabis Cannabinoid Res. 2017 Mar 1;2(1):48-60. doi: 10.1089/can.2016.0037. eCollection 2017. Cannabis Cannabinoid Res. 2017. PMID: 28861504 Free PMC article. Review.
-
Cannabinoid CB1 and CB2 Receptor-Mediated Arrestin Translocation: Species, Subtype, and Agonist-Dependence.Front Pharmacol. 2019 Apr 10;10:350. doi: 10.3389/fphar.2019.00350. eCollection 2019. Front Pharmacol. 2019. PMID: 31024316 Free PMC article.
References
-
- Attramadal H, Arriza JL, Aoki C, Dawson TM, Codina J, Kwatra MM, Snyder SH, Caron MG, Lefkowitz RJ. Beta-arrestin2, a novel member of the arrestin/beta-arrestin gene family. J. Biol. Chem. 1992;267:17882–17890. - PubMed
-
- Gurevich VV, Benovic JL. Visual arrestin binding to rhodopsin. Diverse functional roles of positively charged residues within the phosphorylation-recognition region of arrestin. J. Biol. Chem. 1995;270:6010–6016. - PubMed
-
- Lefkowitz RJ, Inglese J, Koch WJ, Pitcher J, Attramadal H, Caron MG. G-protein-coupled receptors: Regulatory role of receptor kinases and arrestin proteins. Cold Spring Harb. Symp. Quant. Biol. 1992;57:127–133. - PubMed
-
- Lohse MJ, Andexinger S, Pitcher J, Trukawinski S, Codina J, Faure JP, Caron MG, Lefkowitz RJ. Receptor-specific desensitization with purified proteins. Kinase dependence and receptor specificity of beta-arrestin and arrestin in the beta 2-adrenergic receptor and rhodopsin systems. J. Biol. Chem. 1992;267:8558–8564. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

