Pkd1l1 establishes left-right asymmetry and physically interacts with Pkd2
- PMID: 21307093
- PMCID: PMC3042869
- DOI: 10.1242/dev.058149
Pkd1l1 establishes left-right asymmetry and physically interacts with Pkd2
Abstract
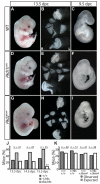
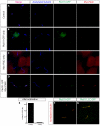
In mammals, left-right (L-R) asymmetry is established by posteriorly oriented cilia driving a leftwards laminar flow in the embryonic node, thereby activating asymmetric gene expression. The two-cilia hypothesis argues that immotile cilia detect and respond to this flow through a Pkd2-mediated mechanism; a putative sensory partner protein has, however, remained unidentified. We have identified the Pkd1-related locus Pkd1l1 as a crucial component of L-R patterning in mouse. Systematic comparison of Pkd1l1 and Pkd2 point mutants reveals strong phenocopying, evidenced by both morphological and molecular markers of sidedness; both mutants fail to activate asymmetric gene expression at the node or in the lateral plate and exhibit right isomerism of the lungs. Node and cilia morphology were normal in mutants and cilia demonstrated typical motility, consistent with Pkd1l1 and Pkd2 activity downstream of nodal flow. Cell biological analysis reveals that Pkd1l1 and Pkd2 localise to the cilium and biochemical experiments demonstrate that they can physically interact. Together with co-expression in the node, these data argue that Pkd1l1 is the elusive Pkd2 binding partner required for L-R patterning and support the two-cilia hypothesis.
Figures





References
-
- Bisgrove B. W., Snarr B. S., Emrazian A., Yost H. J. (2005). Polaris and Polycystin-2 in dorsal forerunner cells and Kupffer's vesicle are required for specification of the zebrafish left-right axis. Dev. Biol. 287, 274-288 - PubMed
-
- Bloodgood R. A. (2010). Sensory reception is an attribute of both primary cilia and motile cilia. J. Cell Sci. 123, 505-509 - PubMed
-
- Blum M., Weber T., Beyer T., Vick P. (2008). Evolution of leftward flow. Semin. Cell Dev. Biol. 20, 464-471 - PubMed
-
- Bogani D., Siggers P., Brixey R., Warr N., Beddow S., Edwards J., Williams D., Wilhelm D., Koopman P., Flavell R. A., et al. (2009). Loss of mitogen-activated protein kinase kinase kinase 4 (MAP3K4) reveals a requirement for MAPK signalling in mouse sex determination. PLoS Biol. 7, e1000196 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

