The role of LAT in increased CD8+ T cell exhaustion in trigeminal ganglia of mice latently infected with herpes simplex virus 1
- PMID: 21307196
- PMCID: PMC3126262
- DOI: 10.1128/JVI.02290-10
The role of LAT in increased CD8+ T cell exhaustion in trigeminal ganglia of mice latently infected with herpes simplex virus 1
Abstract
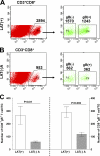
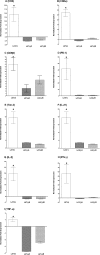
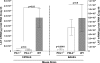
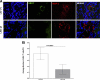
Herpes simplex virus (HSV) infection is a classic example of latent viral infection in humans and experimental animal models. The HSV-1 latency-associated transcript (LAT) plays a major role in the HSV-1 latency reactivation cycle and thus in recurrent disease. Whether the presence of LAT leads to generation of dysfunctional T cell responses in the trigeminal ganglia (TG) of latently infected mice is not known. To address this issue, we used LAT-positive [LAT(+)] and LAT-deficient [LAT(-)] viruses to evaluate the effect of LAT on CD8 T cell exhaustion in TG of latently infected mice. The amount of latency as determined by quantitative reverse transcription-PCR (qRT-PCR) of viral DNA in total TG extracts was 3-fold higher with LAT(+) than with LAT(-) virus. LAT expression and increased latency correlated with increased mRNA levels of CD8, PD-1, and Tim-3. PD-1 is both a marker for exhaustion and a primary factor leading to exhaustion, and Tim-3 can also contribute to exhaustion. These results suggested that LAT(+) TG contain both more CD8(+) T cells and more CD8(+) T cells expressing the exhaustion markers PD-1 and Tim-3. This was confirmed by flow cytometry analyses of expression of CD3/CD8/PD-1/Tim-3, HSV-1, CD8(+) T cell pentamer (specific for a peptide derived from residues 498 to 505 of glycoprotein B [gB(498-505)]), interleukin-2 (IL-2), and tumor necrosis factor alpha (TNF-α). The functional significance of PD-1 and its ligands in HSV-1 latency was demonstrated by the significantly reduced amount of HSV-1 latency in PD-1- and PD-L1-deficient mice. Together, these results may suggest that both PD-1 and Tim-3 are mediators of CD8(+) T cell exhaustion and latency in HSV-1 infection.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

