The pINDUCER lentiviral toolkit for inducible RNA interference in vitro and in vivo
- PMID: 21307310
- PMCID: PMC3048138
- DOI: 10.1073/pnas.1019736108
The pINDUCER lentiviral toolkit for inducible RNA interference in vitro and in vivo
Abstract
The discovery of RNAi has revolutionized loss-of-function genetic studies in mammalian systems. However, significant challenges still remain to fully exploit RNAi for mammalian genetics. For instance, genetic screens and in vivo studies could be broadly improved by methods that allow inducible and uniform gene expression control. To achieve this, we built the lentiviral pINDUCER series of expression vehicles for inducible RNAi in vivo. Using a multicistronic design, pINDUCER vehicles enable tracking of viral transduction and shRNA or cDNA induction in a broad spectrum of mammalian cell types in vivo. They achieve this uniform temporal, dose-dependent, and reversible control of gene expression across heterogenous cell populations via fluorescence-based quantification of reverse tet-transactivator expression. This feature allows isolation of cell populations that exhibit a potent, inducible target knockdown in vitro and in vivo that can be used in human xenotransplantation models to examine cancer drug targets.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


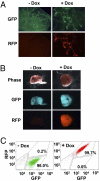
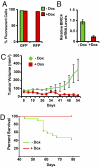
References
-
- Hannon GJ, Rossi JJ. Unlocking the potential of the human genome with RNA interference. Nature. 2004;431:371–378. - PubMed
-
- Elbashir SM, et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. - PubMed
-
- Paddison PJ, et al. A resource for large-scale RNA-interference-based screens in mammals. Nature. 2004;428:427–431. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

