Comparative metagenomics of microbial traits within oceanic viral communities
- PMID: 21307954
- PMCID: PMC3146289
- DOI: 10.1038/ismej.2011.2
Comparative metagenomics of microbial traits within oceanic viral communities
Abstract
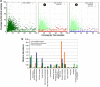
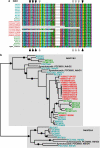
Viral genomes often contain genes recently acquired from microbes. In some cases (for example, psbA) the proteins encoded by these genes have been shown to be important for viral replication. In this study, using a unique search strategy on the Global Ocean Survey (GOS) metagenomes in combination with marine virome and microbiome pyrosequencing-based datasets, we characterize previously undetected microbial metabolic capabilities concealed within the genomes of uncultured marine viral communities. A total of 34 microbial gene families were detected on 452 viral GOS scaffolds. The majority of auxiliary metabolic genes found on these scaffolds have never been reported in phages. Host genes detected in viruses were mainly divided between genes encoding for different energy metabolism pathways, such as electron transport and newly identified photosystem genes, or translation and post-translation mechanism related. Our findings suggest previously undetected ways, in which marine phages adapt to their hosts and improve their fitness, including translation and post-translation level control over the host rather than the already known transcription level control.
Figures


References
-
- Alperovitch A, Sharon I, Rohwer F, Aro E-M, Glaser F, Milo R, et al. Reconstructing a puzzle: existence of cyanophages containing both photosystem-I & photosystem-II gene suites inferred from oceanic matagenomic datasets. Environ Microbiol. 2010;13:24–32. - PubMed
-
- Aono J, Kangawa K, Matsuo H, Horiuchi T. Coliphage 434 tof protein: NH2-terminal amino acid sequence and kinetic and equilibrium measurements of DNA binding. Mol Gen Genet. 1982;186:460–466. - PubMed
-
- Battchikova N, Aro E-M. Cyanobacterial NDH-1 complexes: multiplicity in function and subunit composition. Physiol Plant. 2007;131:22–32. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

